Detection of Pathogenic Leptospiraserovar in Malaysian Wild Rats by Real Time PCR based on LFB1 Gene
Latifah Ibrahim1*, A Abdul Halim1, H Roslina2, M Y Aliza1, H Asmah3, Shafariatul Akmar3, Rahmat MS1 and M Amal Nasir1
1Institute for Medical Research, Jalan Pahang, 50588, Kuala Lumpur, Malaysia
2Clinical Research Centre Sarawak, General Hospital, Kuching, Sarawak, Malaysia
3Universiti Kebangsaan Malaysia, Jalan Muda Abdul Aziz, 50400, Kuala Lumpur, Malaysia
*Address for Correspondence: Latifah Ibrahim, Institute for Medical Research, Kuala Lumpur, Malaysia, E-mail: [email protected]
Submitted: 17 August 2017; Approved: 02 October 2017; Published: 06 October 2017
Citation this article: Ibrahim L, Halim AA, Roslina H, Aliza MY, Picardeau M, et al. Detection of Pathogenic Leptospiraserovar in Malaysian Wild Rats by Real Time PCR based on LFB1 Gene. Int J Vet Sci Technol. 2017;1(2): 035-039.
Copyright: © 2017 Ibrahim L, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Keywords: Laser Capture Micro dissection (LCM); Reference genes; Endometrium; Pyometra
Download Fulltext PDF
Conventional laboratory diagnosis of Leptospirosis is laborious and time consuming. Real time PCR has been reported as cost effective for rapid diagnosis of leptospirosis. In this study, the Real Time PCR method based on LFB1 genes were used to detectpathogenic Leptospira spp in wild rats in selected location in Kuala Lumpur Malaysia (Chow Kit and Dato Keramat market). 140 rats were captured from that area, and samples from kidneys were collected for culture.All the isolates which were obtained were cultured in EMJH supplemented with 3 % rabbit serum. 4 reference genes which are Leptospiracanicola, Leptospirajavanica, Leptospirabataviae and Leptospirahardjo were used to monitor the detection of Leptospira spp. 25 samples were used including one negative sample to detect Leptospirasp. By using Agilent Brilliant III SYBR Master. Mix that consist a novel mutant Taq DNA polymerase which enhance higher nucleotide incorporation and a novel faster-activating hot start method which helps to minimize formation of primer-dimers and increase the specificity of target detection. The primers used were LFB1-F’ and LFB1-R’ for target detection. This gene has been reported to be responsible for the pathogenicity of Leptospira interrogans. All 4 reference gene were successfully amplified and all the 4 reference genes showed the same melting temperature which is 81°C. All amplified samples categorized into two genospecies as two distinct melting curve observed which was 81°C and 83.5°C respectively. In the real time PCR assay a positive reaction was detected by the accumulation of a fluorescent signal calculated in Ct (cycle threshold).All the control pathogenic leptospira produce Ct values ranging from 30.09 to 32.10. Twenty leptospira isolates from the wild rats gave Ct value ranging from 14.27 to 20.22. This study shows that Real Time PCR using the above primer set could be used to identify pathogenic leptospira from local wildrat’s samples.
Introduction
Leptospira is the etiologic agent of leptospirosis, a bacterial zoonosis with worldwide distribution. Leptospirosis is caused by Leptospira sp, a spirochete aerobic bacterium, gram-negative, with spiral morphology [1]. It is an important global disease with public and animal health implications [2]. The number of patients in Malaysia infected with leptospira has increased lately. An outbreak of leptospiral infection was reported among athletes participating in the Eco-Challenge-Sabah 2000 held in Malaysian Borneo. The infection was reported to be associated with water-related activities. Investigation of large outbreaks would be greatly enhanced by the availability of rapid and sensitive diagnostic assays which can confirm the diagnosis early in the clinical illness [3]. 15 cases of leptospirosis and 1 death from secondary school sFtudents in Negeri Sembilan were reported by The Ministry of Health Malaysia in May 2015 [4].
Kuala Lumpur, the capital city of Malaysia, has a large human population and abundance of food supply which attracts rats and makes it a suitable breeding habitat for them [5]. A population of about 6.8 million rats were estimated by Kuala Lumpur City Hall in Kuala Lumpur and this is 4 times the total human population of the city [6].
There are over 230 known serovars in the genus Leptospira. The disease is maintained in nature by chronic renal infection of carrier mammals, which excrete the organism in their urine [7]. A study conducted in 2009 in Malaysia indicated that Leptospira serovar were prevalent in the Malaysian rat population and could be a source of infection to humans. Diagnosis of leptospirosis is usually accomplished retrospectively by serology, because culture requires both special media and incubation for several weeks [8]. Serologic diagnosis by microscopic agglutination test invariably requires testing of acute and convalescent sera, since agglutinating antibodies are not detectable during the acute illness. IgM antibodies become detectable 5 -7 days after the onset of symptoms, and the use of IgM-ELISA assays for presumptive diagnosis has been evaluated in numerous populations [9]. Since leptospira are difficult to culture, several PCR methods have been used to facilitate early diagnosis [10]. A number of PCR assays for leptospiral DNA have been described, but only two have been evaluated in clinical studies used extensively for diagnosis [11]. PCR assays which could detect all pathogenic and non-pathogenic leptospires in clinical samples were also described [12]. More recently, a real-time PCR was developed using Taq Man chemistry (which targeted an 87bp section of the16S rRNA gene of Leptospirasp [13]. We describe here detection of pathogenic leptospira serovar in Malaysian wild rats by Real Time PCR based on LFB1 gene using SYBER Green kit.
Materials and Methods
Sampling
Selection of study area: Two areas in Kuala Lumpur were chosen for the study and the selections were based on suitability of habitat for food, rat breeding and the possibility of spreading the disease. Datuk Keramat (03 ° 09’57.00 “ N, 101 ° 43’52.00 “ E) had an abundance of roadside stalls selling cooked food. Chow Kit (03 ° 09’53.75 “ N, 101 ° 41’56.84 “ E) had the largest fresh food wet market in Kuala Lumpur. The market provides a variety of raw foods, including fruits, vegetables, seafood and meat to the public.
Trapping of rats: The trapping was carried out for 5 days using 100 metal cages measuring 18 cm x 12 cm x 28 cm which were readily available from most hardware shops, and baited with coconut, sweet potatoes or bread. Traps were placed in the evening and were collected early in the morning of the next day, with the assistance of Kuala Lumpur City Hall (DBKL) staff before these areas were crowded with people.
Preparation of EMJH media: About 2.3 g of EMJH Leptospira spp. DifcoTM basic media was dissolved with 900 ml distilled water in a 1 L Scotch bottle at pH 7.5, autoclaved for 15 min, left at room temperature overnight and filtered through a 0.2 µm filter unit. About 100 ml EMJH Leptospira spp. DifcoTM enriched media was added to the 900 ml basic media prepared earlier. Next 5 ml of the media was aliquoted into each of the 10 ml culture tubes and 5-fluorouracil (200 µg/ml) was also added to it. These culture tubes were then kept in 4°C until used.
Sampling techniques: In the laboratory, each of the trapped rats was transferred into a cloth bag and anaesthetised with chloroform in a glass jar for about 30 seconds. Urine samples were collected by direct puncture of the bladder of the rats and 2 drops of each sample placed in culture tubes containing 5 ml EMJH media and 5-fluorouracil (200 µg/ml) prepared earlier. Samples were then incubated for 8 weeks in the dark at 28°C - 30°C. Twenty Leptospira serovar which were isolated from Malaysian wild rats and 4 reference pathogenic strain (Leptospira canicola, Leptospira javanica, Leptospira bataviae and Leptospira hardjo) of Leptospira spp from Unit of Biotechnology, Institute Medical for research, Kuala Lumpur from previous study were used [14]. Leptospira spp was cultured in EMJH medium enriched with 3% fetal calf serum at 28°C and protected from light for 7 days [15].
Identification of pathogenic Leptospira
Microscopic examination: During incubation, samples were observed using dark field microscopy on day 1 until day 3, followed by the first week until the eighth week for the presence of Leptospira spp.
Isolation of Leptospira spp. by culturing: On the 8th week, once the presence of Leptospira spp. was detected, sub-culturing was done by transferring 1.5 ml of the culture into a new 10 ml culture tube containing a 5 ml fresh media of EMJH to obtain a pure culture. Contaminated cultures were filtered with 0.45 µm syringe filter prior to sub culture.
Genomic DNA extraction and Real Time PCR PCR: The culture were washed 3 times, centrifuged at 3000 rpm (10 minutes each time) and processed immediately for DNA extraction. The DNA was extracted using Qiagen DNA Extraction Kit (USA). Primer LFB1-F (50 -CATTCATGTTTCGAATCATTTCAAA-30) and primer LFB1-R (50 -GGCCCAAGTTCCTTCTAAAAG-30) chosen in the locus LA0322 of L. interrogans Lai sequence [10] were used [16]. Prior setting up the reactions, the 2x SYBR Green QPCR master mix and sample was thawed and stored on ice. A master mix was prepared according to the number of reaction of the run. For one reaction preparation required 10 µl of 2× SYBR Green QPCR master mix, 1 µl of upstream primer (adjusted to 500 nM final concentration), 1 µl of downstream primer (adjusted to 500 nM final concentration), 1 µl of DNA template and 7 µl of Nuclease-free water to adjust the final volume to 20 µl. Negative control is replaced by 1 µl of nuclease free water. Each reaction (20 µl) is pipetted into each well of the 8-strips tube and capped with optical strip cap. The reaction is then short-spin to bring all the reagent down to the bottom of the tube and to remove any bubbles. The strips tube was later loaded into Agilent Aria. Mx QPCR thermal cycler for amplification and detection. The QPCR profile run was set to two parts; first part is for amplification and detection while second part is for melt curve analysis. The QPCR profile run start with 95°C for 3 minutes for hot-start , 95°C for 5 seconds for denaturation, and 60°C for 10 seconds for both annealing and elongation. The denaturation and annealing plus elongation were repeated for 40 cycles. After 40 cycles, the temperature was increase to 95°C for denaturation and continue with melt curve profile which start from 65°C and increase 0.5°C for every 5 seconds until it reach 95°C.
Results and Discussion
A rapid, simple and accurate method is needed for the diagnosis of leptospirosis. Direct demonstration of Leptospira clinical samples such as blood, urine, CSF performed by bacterial culture takes a long time and is insensitive [17]. Serum antibody detection, thus, serves as an indirect alternative means of leptospirosis diagnosis. MAT is one of the commonly used antibody detection assays [18]. MAT is insensitive during the early phase of infection and requires a large battery of living Leptospira spp. of various serogroups and serovars which are laborious and costly [17]. Other simpler antibody detection methods have been developed, such as indirect immunoflorescent assay, IgM ELISA and IgM dipstick. Sensitivity of these assays, however, is still limited by the Leptospira spp. used for preparing the antigens. False negative results may occur if the infecting Leptospira spp. does not match the Leptospira spp. used as antigen in the assays. Alternatively, false positive results may be obtained by antibodies in serum of patients who have been infected by previously unrecognized Leptospira infection or exposure to antigenically related organisms especially in the leptospirosis endemic areas [19]. In this study, Real time PCR technique was applied to detect the presence of pathogenic leptospiral DNA in isolates samples of wild rats in selected areas in Chow Kit and Dato Keramat Markets in Kuala Lumpur Malaysia.
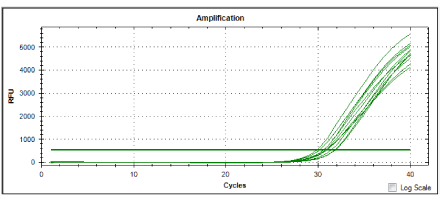
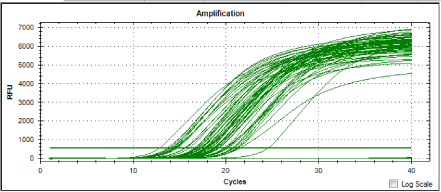
Four pathogenic isolates were used as positive controls. The cultured cultures were. Leptospira interrogans serovar canicola, Leptospira interrogans serovar javanica, Leptospira interrogans serovar bataviae and Leptospira interrogans serovar hardjo. The isolates were obtained from Biotechnology unit, Institute for Medical Research, Kuala Lumpur. figure 1 and figure 2 show amplication plots of 4 reference genes and amplification plot of all samples, respectively. In the real time PCR assay a positive reaction was detected by the accumulation of a fluorescent signal calculated in Ct (cycle threshold): All the control pathogenic leptospiras produced Ct values ranging from 30.09 to 32.10 (Table 1). Twenty leptospira isolates tested from the wild rats gave Ct values ranging from 14.27 to 20.22 (Table 2). In this study, real time PCR technique was applied to detect the pathogenicity of 20 local isolates from wild rats. The local isolates were wild rats from selected market from Chow Kit (S1, S2, S3, S4, S5, S6, S7, S8, S9) and wild rats from selected market from Dato Keramat (S10, S11, S12, S13, S14, S15, S16, S17, S18, S19 and S20) in Malaysia. All the isolates were cultured in EMJH supplemented with 3 % rabbit serum. Real Time PCR with SYBER Green kit was used to amplify the extracted DNA using the set of primers known to amplify the locus LA0322 of Leptospira. This gene is reported to be responsible for the pathogenicity of Leptospira interrogans.
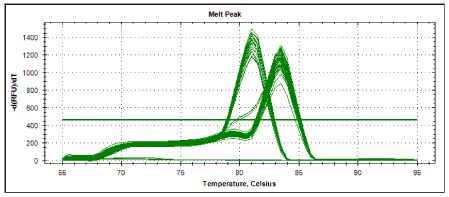
Melting curve analysis showed specific amplifications which allow identification of control pathogenic leptospira (Figure 4). All of the control pathogenic leptospira gave similar melting temperature of 810C. Meanwhile local isolates gave melting temperature either similar to control pathogenic leptospira (Table 4) or a higher melting temperature of 83.50C (Table 5). These enable accurate grouping between the possible pathogenic and non- pathogenic leptospira in local isolates. Therefore, Real Time PCR with SYBR Green as described has the potential as a diagnostic tool in identifying pathogenic leptospira from local wild rats.
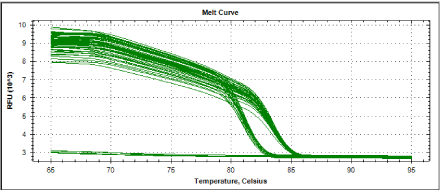
Figure 3 and figure 4 showed melting curves and melting peaks of all samples tested. All amplified samples could seemingly be categorized into two genospecies as two distinct melting curves were observed. One is at 81°C meanwhile the other group showing a melting temperature of 83.5°C. Out of the 20 positive samples, 6 local isolates ( S7, S9, S12, S17, S10 and S15 ) have melting temperature of 81°C and 14 local isolates (S18, S19, S20, S1, S2, S3, S4, S5, S6, S8, S11, S13, S14 and S16 ) have melting temperature of 83.5°C. The leptospira isolates from wild rodents S7, S9, S12, S17, S10 and S15 exhibited melting curves compatible with all the 4 reference strains Leptospira interrogans serovar canicola, Leptospira interrogans serovar javanica, Leptospira interrogans serovar bataviae and Leptospira intern organs serovar hardjo). Therefore, there was a possibility that those isolates were also pathogenic. These data shows the potential of LFB1 real time assay for the detection of pathogenic leptospira from the wild rats. Further research should be done to detect leptospira in animal and human samples using this method
Conclusion
Pathogenic leptospires from the wild rats can be detected using a real time PCR assay based on SYBR Green technology. This method is rapid and simple and could be applied in human as well as animal.
Acknowledgements
We would like to thank the Director General of Health Malaysia for his permission to publish this article. We also thank the Director of Institute for Medical Research for the support and encouragement to publish this paper. This project was financially supported by Ministry of Health research grant (08-CAM-03-02).
- Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA et al. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003; 3: 757-771. https://goo.gl/T5RBLv
- Mc Bride AJ, Athanazio DA, Reis MG. Leptospirosis. Curr Opin Infect. 2005; 18: 681-690. https://goo.gl/QqxTtB
- Sejvar J, Bancroft E, Winthrop K, Bettinger J, Bajani M, Bragg S, et al. Leptospirosis in" eco-challenge" athletes, Malaysian Borneo, 2000. Emerging infectious diseases. 2003; 9: 702-707. https://goo.gl/wVWuv3
- Faine S, Adler B, Bolin C, Perolat, P. Leptospira and Leptospirosis, 2nd edition. Melbourne: MedSci. 1999.
- Levett PN. Leptospira and Leptonema. In: Manual of Clinical Microbiology. Edited by Murray PR, Baron EJ, Jorgensen JH. Washington, DC: American Society for Microbiology. 2003; 929–936.
- Bajani MD, Ashford DA, Bragg SL, Woods CW, Aye T, Spiegel RA et al. Evaluation of four commercially available rapid serologic tests for diagnosis of leptospirosis. J Clin Microbiol.2003; 41: 803-809. https://goo.gl/gy78zm
- Brown PD, Gravekamp C, Carrington DG, Van de Kemp H, Hartskeerl RA, Edwards CN, et al. Evaluation of the polymerase chain reaction for early diagnosis of leptospirosis. J Med Microbiol. 1995; 43: 110-114. https://goo.gl/euzYff
- Merien F, Baranton G, Perolat P. Comparison of polymerase chain reaction with microagglutination test and culture for diagnosis of leptospirosis. J Infect Dis. 1995; 172: 281-285. https://goo.gl/yKJXjB
- Gravekamp C, Van de Kemp H, Franzen M, Carrington D, Schoone G J, Van Eys G J. J. M, et al. Detection of seven species of pathogenic leptospires by PCR using two sets of primers. Journal of General Microbiology. 1993; 139: 1691-1700. https://goo.gl/ZRbqw5
- Smythe LD, Smith IL, Smith GA, Dohnt MF, Symonds ML, Barnett LJ, et al. A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC infectious diseases. 2002; 2:13. https://goo.gl/1pzy8e
- Appassakij H, Silpapojakul K, Wansit R, Woodtayakorn J. Evaluation of the immunofluorescent antibody test for the diagnosis of human leptospirosis. Am J Trop Med Hyg.1995; 52: 340-343. https://goo.gl/kczRco
- Ellinghausen HC. Growth, cultural characteristics, and antibacterial sensitivity of Leptospira interrogansserovar Hardjo. Cornell Vet.1983; 73: 225-239. https://goo.gl/coqFg8
- Levett PN. Leptospirosis. Clin Microbiology Rev.2001; 14 : 296-326. https://goo.gl/D4353T

Sign up for Article Alerts