Analysis of Bacterial Community Structure and Diversity in Different Restoration Methods in Qixing River Wetland
Jihua Wang1*, Yao Ji2, Zening Yuan2, Jianfei Guan2, Chenyu Liu2 and Lixin Lv1
1Harbin Normal University Youth backbone support project (NO.XRQG09)
2Heilongjiang Province postdoctoral research start fund project (LBH-Q13102)
*Address for Correspondence: Jihua Wang, Harbin Normal University Youth backbone support project (NO.XRQG09), E-mail: [email protected] / [email protected]
Submitted: 30 September 2017; Approved: 13 October; Published: 16 October 2017
Citation this article: Wang J, Ji Y, Yuan Z, Guan J, Liu C, et al. Analysis of Bacterial Community Structure and Diversity in Different Restoration Methods in Qixing River Wetland. Adv J Toxicol Curr Res. 2017;1(2): 049-055.
Copyright: © 2017 Wang J, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Download Fulltext PDF
Returning farmland to wetland is a strategic measure for restoration of wetland ecosystems. Bacterial community structure and bacterial diversity in soil are important to the restoration of wetland ecosystems. However, very little research has been conducted to study bacterial communities in natural wetland restoration, particularly the bacterial community composition and diversity of returning farmland to wetland. We used high-throughput sequencing technology to compare the bacterial community structure and diversity of Natural Wetland (NW) soil with Returning Farmland to Wetland (RFW) and Returning Farmland to Forest (RFF) soils in Qixing River wetland. The following phyla were present in all samples: Proteobacteria, Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Verrucomicrobia, Gemmatimonadetes, Nitrospira, Planctomycetes, Cyanobacteria, and Saccharibacteria. Of these, Proteobacteria (20.8–32.6%) Acidobacteria (14.4–33.9%) and Actinobacteria (11.4–19.0%) had the highest relative abundance in all the soil samples. The relative abundance values of Proteobacteria, Acidobacteria, and Actinobacteria were similar between RFW and NW soils. In addition, study of alpha diversity indices indicated that the ACE, Chao, and Shannon indices were significantly higher in RFW soils than NW and RFF soils. Redundancy Analysis (RDA) revealed that the total salt, organic matter, and total nitrogen significantly affected the bacterial community composition, while the total salt, organic matter, and total nitrogen were the dominant environmental factors in RFW soils. The present research indicated that RFW improved bacterial community diversity and the total salt, organic matter, and total nitrogen contents.
Introduction
Wetlands exist in the transitional zone between terrestrial and aquatic environments, and play important roles in water storage and treatment, global climate regulation, and wildlife habitat [1]. Studies have shown that plants, animals, as well as microorganisms are abundant in wetland ecosystems [2].Microbes play the role of decomposers in these ecosystem and their primary function is energy flow and substance transformation. Meanwhile, microorganisms influence the succession and differentiation of wetland ecosystems [3]. Previous studies have demonstrated that bacterial communities constitute the majority of microorganisms in wetland habitats, [4]. Dynamic changes in the bacterial community point to changes in the wetland ecosystem environment and directly correspond to changes in soil health [5]. Studying the change of soil microorganisms under wetland ecological restoration are beneficial to realize the stage of wetland restoration [6]. A previous study in Yellow River wetland indicated that bacteria represent the largest group of microorganisms inhabiting the soil, accounting for 62.87–96.64% of the total population of soil microorganisms after returning farmland to wetland [7]. Environmental factors such as the total salt, pH, and soil water content have been reported to influence bacterial growth, which is very sensitive to changes in the soil quality [8]. It is essential to research the relationship between the bacterial community structure and environmental changes [9].
Following recent advances in sequencing technology, high-throughput sequencing has been widely used to study community structures of bacteria in soil, sediments, and freshwater [10-12]. Wang et al. reported that total organic matter, total phosphorus, salinity, and total nitrogen significantly influenced bacterial community structure under different soil environments [13]. Bolhius and Stal studied bacterial community composition in coastal microbial mats via 16S rRNA gene tag sequencing technology and reported that bacterial diversity is influenced by seasonal variation and salinity via [14]. Furthermore, Proteobacteria, Bacteroidetes, and Firmicutes were the dominant phylum in the Winogradsky columns by testing bacterial diversity using high-throughput sequencing technology [15].
The Qixing River wetland is a freshwater marsh in the Sanjiang Plain, in Northeast China. Qixing River wetland has been listed in the Ramsar Convention in 2011 as a wetland of international importance [16]. The wetland covers about 2 × 104 ha and it is an important component of the wetland ecosystem in the cold temperature zone [16]. The Qixing River wetland is categorized into three types, including Returning Farmland to Wetland (RFW), Natural Wetland (NW), and Returning Farmland to Forest (RFF). RFW is a strategic measure for wetland ecological restoration [17]. However, no studies have investigated the bacterial community structure in Qixing River wetland. Therefore, in the present study we aimed to study the correlation between bacterial community composition and environmental factors during the restoration of Qixing River wetland using high-throughput sequencing technology. Through this research, we hope to shed insight into the restoration process of returning farmland to wetland, which could serve as a scientific basis for returning farmland to wetland and help in the scientific management of wetland restoration in the Sanjiang Plain.
Materials and Methods
Study area description
Qixing River wetland is located in Sanjiang Plain, Heilongjiang Province, China (46°40'–46°52' N, 132°05'–132°26' E). The wetland includes regions with ongoing Returning Farmland to Wetland (RFW), Natural Wetland (NW), and Returning Farmland to Forest (RFF) projects. It has a semi-humid continental monsoon climate with a mean annual air temperature is 2.35' [18] and average annual rainfall of 551.5 mm. The Qixing River wetland has different soil types, primarily consisting of pulp soil and swamp soil [19]. Phragmites australis and Calamagrostis angustifolia are the dominant plant species in the wetland.
Sample collection
In the RFW region of Qixing River wetland (46°43'10.8' N, 132°11'27.3' E), samples were collected in July 2014 (RFW1), October 2014 (RFW2), and May 2015 (RFW3) three times. C. angustifolia is the dominant plant species in RFW. According to the previous studies, the bacterial diversity was the highest in autumn [20]. Therefore, in order to compare the bacterial community composition between the NW (46°43'03.0” N, 132°11'23.7” E) and RFF (46°42'20.2” N, 132°06'03.4” E) regions, we collected soil samples from these two areas in October 2014. The dominant plant species in NW was P. australis and the main plant species in RFF was Populus L. There were three RFW, NW, and RFF plots, and three replicates were carried out for each plot. All soil samples were collected at a soil depth of 0–20 cm. The soil samples collected from each plot were thoroughly mixed, and stored in aseptic bags at 0–4' during transport to the laboratory. In the laboratory, one part of the samples was dried for testing soil characteristics and the other part was stored at –80' until analysis of bacterial community structure.
Analysis of soil physicochemical properties
To measure the soil Water Content (WC), soil samples were dried at 105' for 24 h. Soil pH was determined using a compound electrode with a soil to water ratio of 1:2.5. Soil Organic Matter (OM) was determined using the potassium dichromate oxidation-outer heating method. Soil Total Nitrogen (TN) was measured by Kjeldahl digestion. Soil Total Phosphorus (TP) was measured using molybdenum antimony colorimetry. Soil Total Salt (TS) was determined using quality method.
DNA extraction and PCR amplification of soil bacteria
Bacterial DNA was extracted from the samples of Qixing River wetland by using the E.Z.N.A. ® soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to manufacturer’s protocols. The V4-V5 region of the bacteria 16S ribosomal RNA gene were amplified by PCR (95' for 3 min, followed by 27 cycles at 95' for 30 s, 55' for 30 s, and 72' for 45 s and a final extension at 72' for 10 min) using primers 338F (5’-barcode- ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’), where barcode is an eight-base sequence unique to each sample. PCR reactions were performed in triplicate 20 μL mixture containing 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA.
Illumina MiSeq sequencing and accession number
Amplicons were extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, U.S.) according to the manufacturer’s instructions and quantified using QuantiFluor™ -ST (Promega, U.S.). Purified amplicons were pooled in equimolar and paired-end sequenced (2×250) on an Illumina MiSeq platform according to the standard protocols. The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP******).
Bioinformatics and statistical analyses
Raw FASTQ files were demultiplexed and quality-filtered using QIIME (version 1.9.1) using the following criteria: (i) 300-bp reads were truncated at any site with an average quality score of < 20 over a 50-bp sliding window, discarding truncated reads shorter than 50 bp; (ii) exact barcode matching, two nucleotide mismatch in primer matching, and reads containing ambiguous characters were removed, and (iii) sequences with overlaps of more than 10 bp were assembled according to their overlap sequence. Reads that could not be assembled were discarded.
Operational Taxonomic Units (OTUs) were clustered with 97% similarity cutoff using UPARSE (version 7.1 http://drive5.com/uparse/) [21], and chimeric sequences were identified and removed using UCHIME. The taxonomy of each 16S rRNA gene sequence was analyzed by RDP Classifier (http://rdp.cme.msu.edu/) against the Silva (SSU115) 16S rRNA database using a confidence threshold of 70%.
Canoco 4.5 was used to identify correlation between environmental factors and bacterial community structure, and the figure was generated by CanoDrao 4.0. Data in tables were analyzed by SPSS 22.0 software. Values in the tables indicate the means of the samples.
Results
Physicochemical characterization of sample sites
In Qixing River wetland, the soil nutrient content was higher in the RFW than in the RFF and NW samples, especially in the October 2014 RFW sample (RFW2). The OM, TN, TP, and TS contents were higher in RFW2 than in NW and RFF (Table 1). The highest and lowest OM values were 17.50% and 9.38% and were found in RFW2 and RFF, respectively. The TS values ranged from 0.57 g/kg in RFF to 2.07 g/kg in RFW2. The TN increased from 0.35% in RFF to 0.73% in RFW2. The TP values increased from 0.067% in RFF to 0.154% in RFW2. The WC ranged from 22.30% in RFF to 44.30% in NW. However, the pH values of all the samples were nearly neutral (6.80–7.30).
Bacterial alpha diversity indices
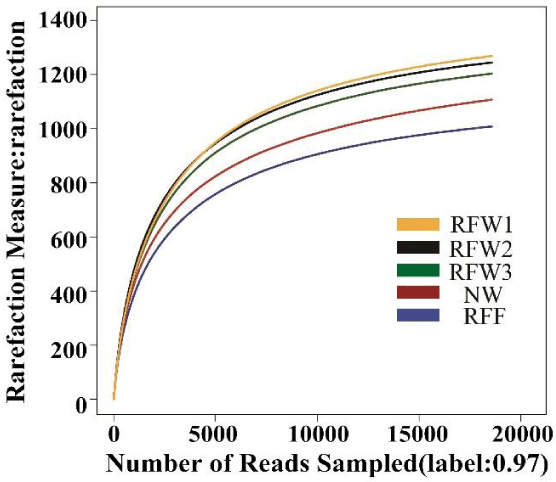
Figure 1 presents a rarefaction curve of bacterial community at similarity level of 0.97. The OTU curve of the five samples tended to plateau when the sequence numbers reached 18567. The current results showed that the samples were reasonable and the sequencing depth covered all species in the five samples.
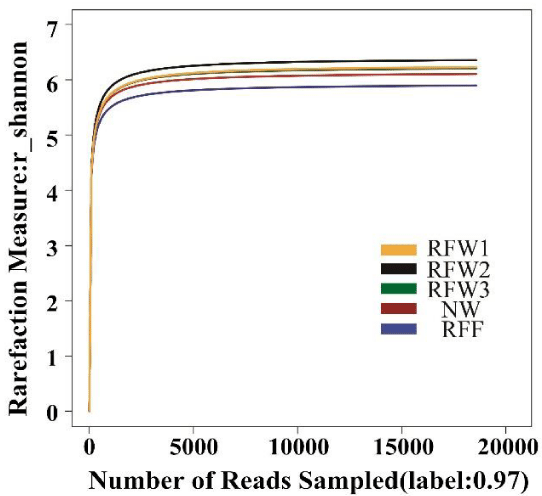
A total of 1, 85, 601 sequences were obtained from the five samples by high-throughput sequencing technology. A similarity level of 97% was used to identify OTUs and to estimate diversity (Figure 2, Table 2). The results revealed that coverage was higher than 0.99 in all the samples. Therefore, the results of the high-throughput sequencing could adequately represent the bacterial community structure in Qixing River wetland (Table 2). As shown in figure 2, the Shannon diversity index was the highest in RFW2 and the lowest in RFF (Figure 2).
The ACE estimate of bacterial diversity ranged from 1087 to 1356 and the Chao estimate ranged from 1102 to 1380 (Table 2). The ACE and Chao values were significantly higher in RFW2 than in NW and RFF. The Shannon diversity estimate ranged from 5.89 in RFF to 6.36 in RFW2. The Simpson diversity estimate ranged from 0.0032 in RFW2 to 0.0059 in RFF. The Shannon and Simpson indices were significantly higher in RFW2 than in NW and RFF (Table 2).
Phylum-level taxonomic distribution
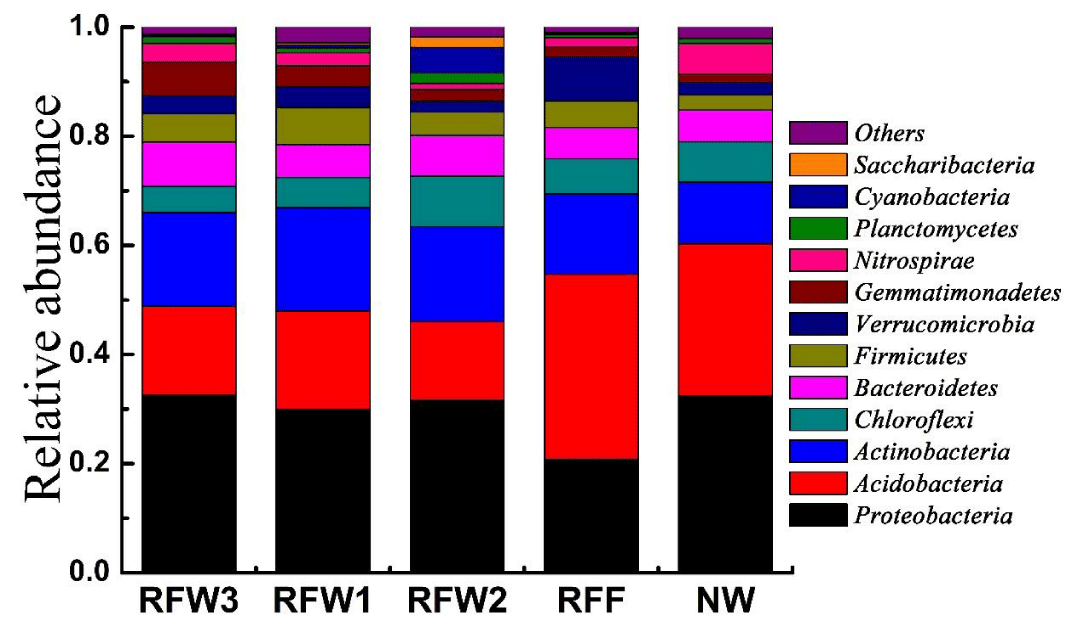
As shown in figure 3, based on the results of the high-throughput sequencing technology, the bacterial species in in RFW1, RFW2, RFW3, NW, and RFF belonged to 12 phyla, including Proteobacteria, Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Verrucomicrobia, Gemmatimonadetes, Nitrospiras, Planctomycetes, Cyanobacteria, and Saccharibacteria were included in five samples. Proteobacteria, Acidobacteria, and Actinobacteria. Of these, Proteobacteria had the highest relative abundance in RFW1, RFW2, RFW3, and NW, ranging from 29.9% to 32.6%. Acidobacteria was the highest abundance in RFF with a relative abundance of 33.9%. The relative abundance of Actinobacteria ranged from 11.4% to 19.0%.
Genus-level distribution
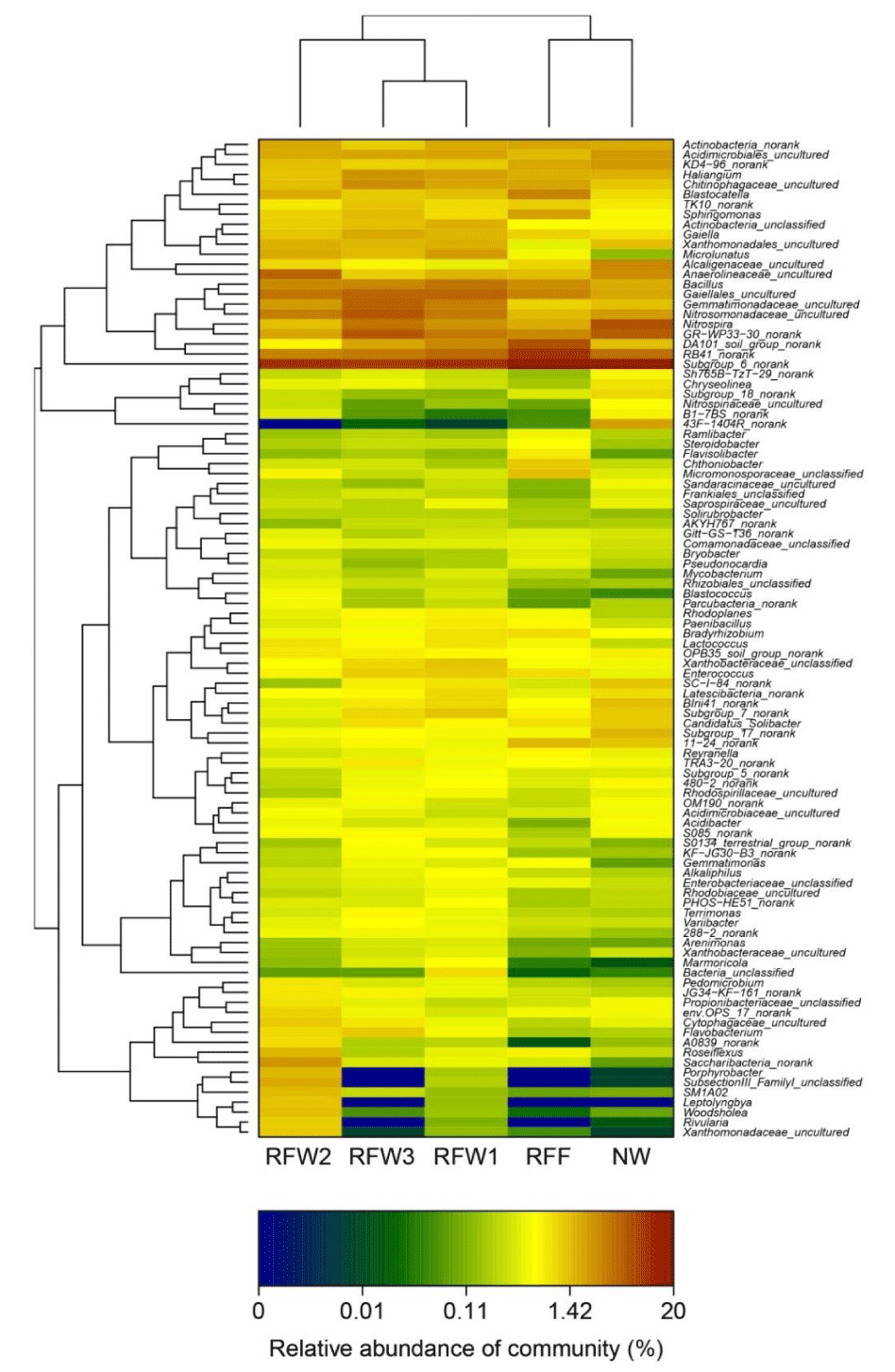
As shown in figure 4, the relative abundance of Subgroup_6_norank was the highest in all these zones, ranging from 8.5% to 20%. Furthermore, Anaerolineaceae_uncultured was relatively abundant in RFW2; the relative abundance values of GR-WP33-30_norank, Gemmatimonadaceae_uncultured, and Gaiellales_uncultured were higher in RFW3; those of RB41_norank and DA101_soil_group_norank were higher in RFF; and those of GR-WP33-30_norank and Nitrospira were over 5% in NW.
In addition, as shown in figure 4 (from Actinobacteria_norank to Subgroup_6_norank), the relative abundance values of Anaerolineaceae_uncultured and Xanthomonadales_uncultured were higher in RFW2 than in the other sampling groups. Additionally, from Sh765B-TzT-29_norank to Bacteria_unclassified (Figure 4, Acidimicrobiaceae_uncultured, OPB35_soil_group_norank, Parcubacteria_norank, Blastococcus, Rhizobiales_unclassified, Comamonadaceae_unclassified, and Gitt-GS-136_norank all had relatively higher abundance in RFW2 than in RFWI, FRW3, NW, and RFF. From Pedomicrobium to Xanthomonadaceae_uncultured (Figure 4), the relative abundance values of Xanthomonadaceae_uncultured, Rivularia, Woodsholea, Leptolyngbya, SM1A02, SubsectionIII_FamilyI_unclassified, Porphyrobacter, Saccharibacteria_norank, Roseiflexus, A0839_norank, Flavobacterium, Cytophagaceae_uncultured, env.OPS_17_norank, Propionibacteriaceae_unclassified, JG34-KF-161_norank, and Pedomicrobium were significantly higher in RFW2 than in the other zones.
Relationship between bacterial community composition and environmental factors
We used correlation analysis to investigate the relationship between bacterial diversity and environmental factors (Table 3). As shown in the table, there were significantly positive correlations between TP, TS and Shannon indices (P < 0.01) and between OM, TN, and Shannon (P < 0.05; Table 3). There were significantly positive correlations between ACE and TP (P < 0.05). However, negative correlations were observed between ACE, Shannon, and pH (pH value: 6.8–7.3). The results suggested that high nutrient content benefited bacterial growth in the Qixing River wetland and contributed to improve diversity in the bacterial community.
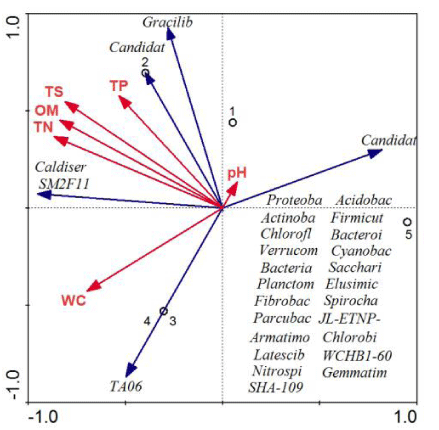
Figure 5 illustrates the relationships between the environmental factors and the sample plots. RDA was used to determine the correlation between the bacterial community structure and environmental factors. Six environmental factors were selected and included in the RDA (Figure 5). Among the environmental factors influencing the bacterial community composition, TS, OM, and TN were the dominant environmental factors in this study. TS, OM, and TN significantly influenced the bacterial groups in RFW2 (Figure 5, Table 4). However, these factors only slightly influenced the bacterial community structure in RFF (Figure 5). At the phylum level, SM2F11 and Caldiserica as well as Candidate_division_WS6 and Gracilibacteria had little differences in Qixing River wetland (Figure 5).
Discussions
A previous study reported that estimation of the alpha diversity, which reflects species diversity of the single sample, is an effective method to analyze bacterial group composition (Paul FK and Josephine Y A, 2004) Meanwhile, alpha diversity reacted bacterial community richness and diversity, including diversity estimators (ACE and Chao) as well as richness estimators (Shannon and Simpson) [22,23]. Higher values of ACE and Chao indices indicate greater diversity within the bacterial community. Higher values of the Shannon index indicates greater richness of the bacterial community, while a lower Simpson index represents greater richness. In the present research, we investigated bacterial diversity during the restoration process of the RFW, NW, and RFF zones in Qixing River wetland. The results revealed significant differences in bacterial diversity, especially in the ACE, Chao, and Shannon indices in RFW2, which were significantly higher than the corresponding values in NW and RFF. On the other hand, the Simpson index was lower in RFW2 than in NW and RFF (Table 2). Together, these results demonstrated that bacterial diversity was the highest in RFW, indicating that returning farmland to wetland was a reasonable approach for wetland restoration. The results of the present study revealed that RFW contributed to wetland reconstruction during the restoration of Qixing River wetland. A previous study found that higher bacterial diversity could be related to vegetation type and physicochemical characteristics [24].
The results of the high-throughput sequencing revealed that Proteobacteria, Acidobacteria, and Actinobacteria were the dominant phyla in RFW1, RFW2, RFW3, NW, and RFF (Figure 3). These results are consistent with those of the previous study [27]. Proteobacteria (29.9–32.6%) exhibited the highest abundance in RFW1, RFW2, RFW3, and NW. Studies have already reported that Proteobacteria survive not only in acidic soil in the natural wetland of Virginia [25] but also in alkaline soil in the inland wetland of La Brava [26]. The soil pH of Qixing River wetland was found to be nearly neutral. Therefore, Proteobacteria was widely distributed in this habitat, which is similar to the findings reported by Sun et al. [11]. Generally, Acidobacteria is a highly abundant and diverse phylum of the domain Bacteria [27,28], and it accounts for 20–46% of the total bacteria in soil [27,29]. Based on our study, Acidobacteria (33.9%) had the highest relative abundance among all the phyla in RFF. The distribution of Acidobacteria in soil was decreased with increasing soil moisture content [30]. Actinobacteria has been previously detected as the main phylum in wetland soils, with a relative abundance exceeding 10% [31]. The relative abundance values of Actinobacteria were higher in RFW than in the other wetland zones NW and RFF, indicating that the environmental conditions of RFW contributed supported the distribution of Actinobacteria. The relative abundance values of Proteobacteria, Acidobacteria, and Actinobacteria in RFW1, RFW2, and RFW3 were similar to the corresponding values in NW (Figure 3). The results indicated that returning farmland to wetland was the most efficient restoration project in Qixing River wetland. Furthermore, as RFF did not recover at the same level as RFW, RFF may require a longer time to achieve NW status.
The bacterial community structure and bacterial diversity were both affected by environmental factors in Qixing River wetland (Figure 5, Table 3). As shown by the results of the RDA, the TS, OM, and TN were the main environmental factors influencing the structure and diversity of the bacterial community in the restoration process of Qixing River wetland. Previous studies have reported that the bacterial community composition is primarily affected by TS and TN at the global scale [31,32]. The TS, OM, and TN values were higher in RFW than in NW and RFF (Table 1), suggesting that returning farmland to wetland was the most effective method to achieve restoration in Qixing River wetland. Among the soil environmental factors, TS, OM, and TN were found to be the most important contributors to bacterial community in RFW2. However, TS, OM, and TN only slightly influenced the bacterial community structure in RFF. This may be because additional environmental factors (such as plant species and other uncertain environment variables) may contribute to the differences in bacterial community composition in RFF [33]. Therefore, returning farmland to wetland is the most effective reconstruction strategy for Qixing River wetland.
Conclusion
Here, we used high-throughput sequencing technology to investigate bacterial communities in areas of the Qixing River wetland undergoing RFW, NW, and RFF projects. At the phylum level, Proteobacteria, Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Verrucomicrobia, Gemmatimonadetes, Nitrospiras, Planctomycetes, Cyanobacteria, and Saccharibacteria were present in the RFW1, RFW2, RFW3, NW, and RFF zones. Of these, Proteobacteria, Acidobacteria, and Actinobacteria were the dominant phyla in all the samples. The ACE and Chao indices were higher in RFW than in NW and RFF. The Shannon index was significantly higher in RFW than in NW and RFF. Correlation analysis revealed a significantly positive correlation between TS, TP and the Shannon index (P < 0.01). Results of the RDA suggested that TS, OM, and TN significantly affected bacterial community composition in Qixing River wetland, especially in RFW2. Our results indicated that RFW is an effective method for restoration of Qixing River wetland. However, long-term studies are needed to verify the efficacy of restoration of RFF.
- He S, Malfatti SA, McFarland JW, Anderson FE, Pati A, Huntemann M, et al. Patterns in wetland microbial community composition and functional gene repertoire associated with methane emissions. MBio. 2015; 6: 66-15. https://goo.gl/KfTYaU
- Pei XC, Xu YL, Wei w. A review on soil microorganisms wetland ecosystem. Wetland Science. 2009;7: 181-186. https://goo.gl/3L2qZb
- Chen H H, Xu XH, Li HT. Diversity analysis of soil fungal community structure in Sanjiang wetland by PCR-DGGE. Research of Environmental Sciences. 2012; 25: 1272-1278. https://goo.gl/E4Y6i7
- Duguma D, Rugman-Jones P, Kaufman MG, Hall MW, Neufeld JD, Stouthamer R, et al. Bacterial communities associated with culex mosquito larvae and two emergent aquatic plants of bioremediation importance. PLoS ONE. 2013; 8: 72522. https://goo.gl/9E1gN7
- Wang J L, Liu J Z, Chen Z L, Kuang Y B. Effects of enrofloxacin residues on the functions of soil microbes. Acta Ecologica Sinica. 2005; 25: 279-282. https://goo.gl/fGEbev
- Meng XD, Zhang PJ, Li ZX. A review on soil on soil microorganism under wetland restoration. YuNan Geographic Environment Research. 2011; 23: 101-105. https://goo.gl/FfHzky
- Zhu N, Zhao TQ, Xiao CY, Jiao LH, Zhang KF. Distribution characters of soil microbial population in the restored riparian wetland. Journal of Henan polytechnic university. 2012; 31: 352-356. https://goo.gl/JA1CED
- Lin X G, Yin R, Zhang H Y, Huang JF, Chen RR, Cao ZH. Changes of soil microbiological properties caused by land use changing from rice-wheat rotation to vegetable cultivation.Emvironmental Geochemistry and Health. 2006; 26: 119-128. https://goo.gl/dQsxq6
- Lipson D A, Schadt C W. Changes in soil microbial community structure and function in an alpine dry meadow following spring snowmelt. Microbial Ecology. 2002; 43: 307-314. https://goo.gl/FK2dk5
- Schmidt, P A, Bálint M, Greshake B, Bandow C, Römbke J, et al. Illumina metabarcoding of a soil fungal community. Soil Biol Biochem. 2013; 65: 128–132. https://goo.gl/wy8guh
- Sun W, Xiao T, Sun M, Dong Y, Ning Z, Xiao E, et al. Diversity of the sediment microbial community in the Aha watershed (Southwest China) in response to acid mine drainage pollution gradients. Appl Environ Microbiol. 2015;81: 4874-4884. https://goo.gl/s8GTy9
- Zhang H H, Huang T L. Archaeal community structure and quantity in the oxygen deficient sediments from three water supply reservoirs.J Pure Appl. Microb. 2013; 7: 2783–2789. https://goo.gl/hs5kBn
- Wang Y, Sheng H F, He Y, Wu J Y, Jiang Y X, Tam N F Y, et al. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl Environ Microb. 2012; 23, 8264–8271. https://goo.gl/jztqcu
- Henk Bolhuis, Lucas J Stal. Analysis of bacterial and archaeal diversity in coastal microbial mats using massive parallel 16S rRNA gene tag sequencing. International Society for Microbial Ecology. 2011; 5: 1701-1712. https://goo.gl/hG3mKu
- Rundell EA, Banta LM, Ward DV, Watts CD, Birren B, Esteban DJ. 16S rRNA Gene Survey of Microbial Communities in Winogradsky Columns.PLoS ONE. 2014; 9: 104134. https://goo.gl/iADu1C
- Liu ZY, Wang H, Huo LY, Cui SB, Yuan ZN. Mechanism of ecological compensation of wetland protection in Songhua River Basin. Wetland Science. 2015; 13: 202-206. https://goo.gl/NJz9pM
- Zhao YQ, Zhang PJ. Ecological Restoration of soils in reclaimed wetland from cultivation. Wetland science & management. 2010; 6: 55-58. https://goo.gl/epCLCg
- Wang F, Gao YG, Bai MQ. Impact of climate change on natural vegetation net primary productivity in Qixing River Wetland ecosystem from 1961-2008. Chinese Agricultural Science Bulletin. 2011; 27: 257-262. https://goo.gl/dLwjcB
- Li L, Chen CH, Gao DW. Diversity and abundance of ammonia-oxidizing archaea and its correlation with environmental factors in Qixing River Wetland. Acta Scientiae Circumstantiae. 2015; 35: 1097-1105. https://goo.gl/bPxBhV
- Zeng XL, Liu XG, Wu ZF, Shi X, Lu SM. Community characteristics of ANAMMOX bacteria in subsurface flow constructed wetland (SSFCW) for processing of aquaculture waste water. Environmental Science. 2016; 37: 615-621. https://goo.gl/jkzPQD
- Edgar RC. UPARSE highly accurate OUT sequences from microbial amplicon reads. Nat Methods. 2013; 10: 996-998. https://goo.gl/GPqTMf
- Lv XF, Yu JB, Fu YQ, Ma B, Qu FZ, Ning K, et al. 2014. A meta-analysis of the bacterial and archaeal diversity observed in wetland soils. The Scientific World Journal. 2014; 2014: 437684. https://goo.gl/Ni7MhV
- Zhang HH, Chen SN, Huang TL, Ma WX, Xu JL, Sun X. Vertical distribution of bacterial community diversity and water quality during the reservoir thermal stratification. Int J Environ Res Public Health. 2015; 12: 6933-6945. https://goo.gl/FRDq3i
- Stottmeister U, Wiebner A, Kuschk P, Kappelmeyer U, Kastner M, Bederski O et al. Effects of plants and microorganisms in constructed wetlands for wastewater treatment. Biotechnology Advances. 2003;22: 93-117. https://goo.gl/6o49UT
- Peralta RM, Ahn C, Gillevet PM. Characterization of soil bacterial community structure and physicochemical properties in created and natural wetlands. The Science of the Total Environment. 2013; 443: 725-732. https://goo.gl/mbYVgz
- Farias ME, Contreras M, Rasuk MC, Kurth D, Flores MR, Poire DG et al. Characterization of bacterial diversity associated with microbial mats, gypsum evaporites and carbonate microbialites in thalassic wetlands: Tebenquiche and La Brava, Salar de Atacama, Chile. Extremophiles. 2014;18: 311-329. https://goo.gl/9ZchnQ
- Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16s rRNA genes. Appl Environ Microbiol. 2006;72: 1719-1728. https://goo.gl/C45F2H
- Kraigher B, Stres B, Hacin J, Ausec L, Mahne I, Elsas JD, et al. Microbial activity and community structure in two drained fen soils in the Ljubljana Marsh.Soil Biol Biochem. 2006; 38: 2762-2771. https://goo.gl/K2rrUX
- Lee SH, Ka JO, Cho JC. Members of the phylum Acidobacteria a dominant and metabolically active in rhizosphere soil.FEM Microbiology Letter. 2008; 285: 263-269. https://goo.gl/nXGruJ
- Sui X, Zhang RT, Zhong HX, Xu N, Wang JF, Liu YZ, et al. Study on Bacterial Diversity of Deyeuxia angustifolia wetland by Application of High-throughput Sequencing Technology in Sanjiang Plain. Soils. 2015; 47: 919-925. https://goo.gl/ZDfYRy
- Deng Y, Cui X, Hernandez M, Dumont MG. Microbial Diversity in Hummock and Hollow Solis of Three Wetlands on the Qinghai-Tibetan Plateau Revealed by 16S rRNA Pyrosequencing. PLoS ONE. 2014; 9: e103115. https://goo.gl/4kb27k
- Morrissey EM, Franklin RB. Evolutionary history influences the salinity preference of bacterial taxa in wetland soils. Front Microbiol. 2015; 6: 1013. https://goo.gl/jgW98m
- Zhang W, Wu XK, Liu GX, Chen T, Zhang GS, Dong ZB, et al. Pyrosequencing reveals bacterial diversity in the rhizosphere of three Phragmites australis ecotypes. Geomicrobiology. 2013; 30: 593-599. https://goo.gl/ergWak

Sign up for Article Alerts