Molecular and Epigenetic Mechanisms of Bidirectional Liver Fibrosis?
Krishna Sumanth Nallagangula1, Ramesh Pradhan2 and Shashidhar.K.N1*
1Department of Biochemistry, Sri Devaraj Urs Medical College, SDUAHER, Tamaka, Kolar, Karnataka, India
2Department of Biochemistry, GCS Medical College, Hospital & Research Centre, Ahmedabad, Gujarat, India
*Address for Correspondence: Shashidhar KN, Professor & Head of Department, Department of Biochemistry, Sri Devaraj Urs Medical College, SDUAHER, Tamaka, Kolar, Karnataka, India, Tel: +91 9845248742; E-mail: drshashikn1971@yahoo.co.in
Submitted: 12 December 2017; Approved: 26 December 2017; Published: 27 December 2017
Citation this article: Putnam C. Molecular and Epigenetic Mechanisms of Bidirectional Liver Fibrosis. Int J Ophthal Vision Res. 2017;3(1): 001-002.
Copyright: Sumanth NK, Pradhan R, Shashidhar KN. Molecular and Epigenetic Mechanisms of Bidirectional Liver Fibrosis. American J Liver Clinical Res. 2017;1(1):001-006.
Keywords: Liver fibrosis; Hepatic regeneration; Cytokines; Epigenetics
Download Fulltext PDF
Liver fibrosis is natural wound healing response to different etiologies of chronic liver insults leading to accumulation of Extra Cellular Matrix (ECM) due to imbalance between synthesis and degradation. Fibrogenesis is consequence of multicellular response; activation of Hepatic Stellate Cells (HSCs) and transdifferentiation into myofibroblasts are crucial for development of hepatic scar. Recent studies evidenced that liver fibrosis is potentially bidirectional regulated by complex cytokines, growth factors, genetic and epigenetic mechanisms (DNA methylation, histone modifications and miRNAs mediated gene silencing). Regression of liver fibrosis is due to increase in collagenolytic activity and increased Metalloproteinase (MMPs) activity with decreased expression and activity of Tissue Inhibitors of Metalloproteinases (TIMPs). Reversible epigenetic mechanisms, pro-fibrotic and anti-fibrotic miRNAs regulate progression and regression of liver fibrosis which initiates to discover diagnostic, prognostic and therapeutic should be comprehensively defined. Hence, in this review we made an attempt to understand molecular, genetic and epigenetic mechanisms of bidirectional liver fibrosis.
Abbreviations
ALT: Alanine aminotransferase; AST: Aspartate aminotransferase; GLDH: Glutamate Dehydrogenase; γGT: Gamma Glutamyl Transferase; LDH: Lactate Dehydrogenase; CRP: C-Reactive Protein; α2M: α 2 Macroglobulin; CTGF: Connective Tissue Growth Factor; PIIINP: Procollagen III amino peptide; MMPs: Matrix Metallo Proteinases; TIMPs: Tissue Inhibitors of Metalloproteinases; HBV: Hepatitis B Virus; HCV: Hepatitis C Virus; ASH: Alcoholic Steatohepatitis; NAFLD: Non Alcoholic Fatty Liver Diseases; IGF-1: Insulin like Growth Factor 1; EMT: Epithelial Mesenchymal Transition; TGF-β: Transforming Growth Factor β; PDGF: Platelet Derived Growth Factor; ET-1: Endothelin-1; ROS: Reactive Oxygen Species
Introduction
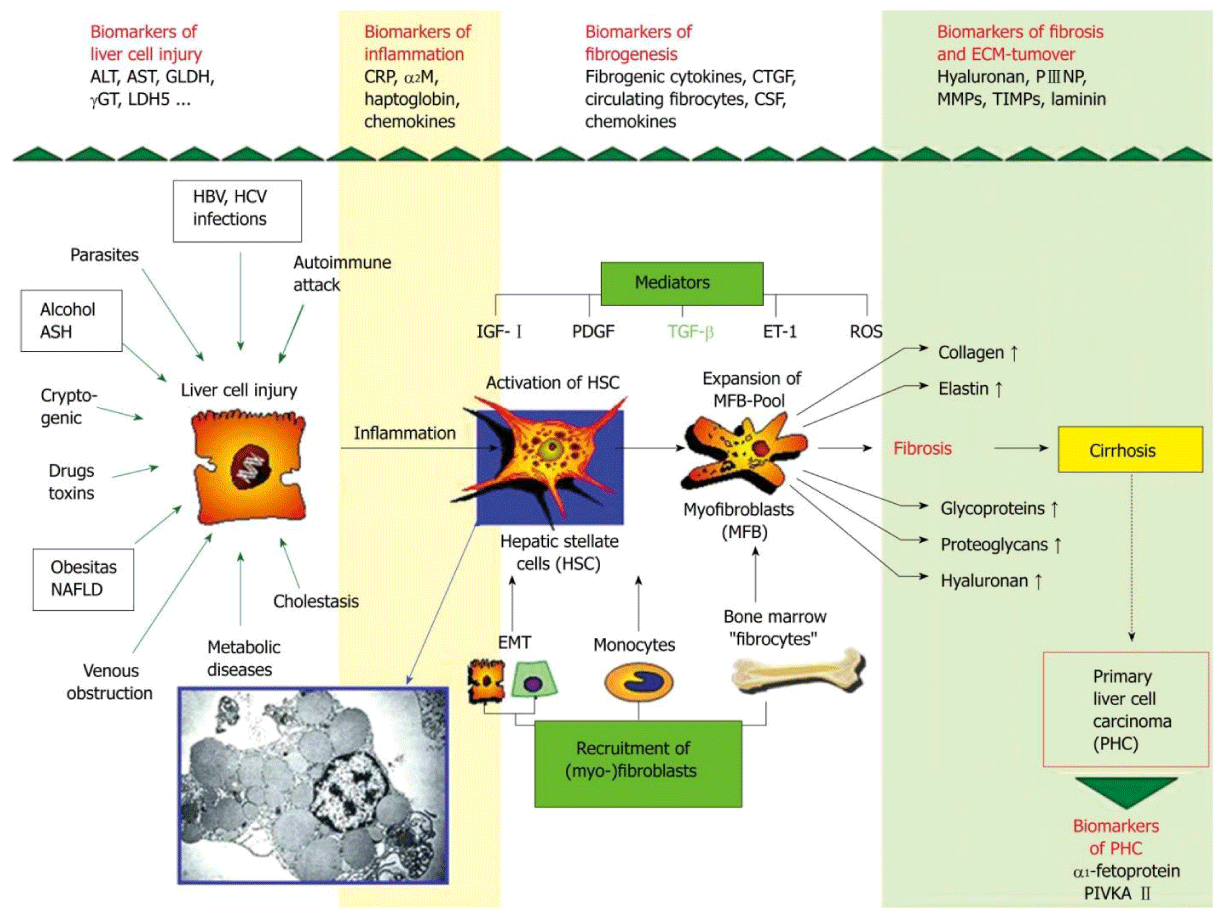
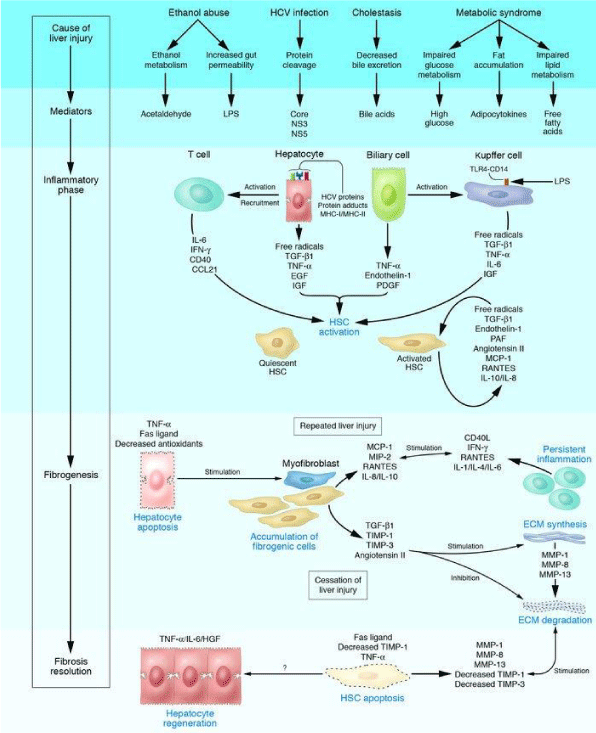
Liver, a vital organ performs several crucial functions; substrate metabolism, detoxification, protein and digestive enzyme synthesis and immune response for human survival [1]. Being highly vascular organ, it is continuously exposed to injury and damage by hepatotoxins viz viruses, drugs, alcohol, excess fat etc., leading to inflammation and fibrosis [2]. Liver fibrosis is natural wound healing response to chronic liver insults which involves deposition of Extra Cellular Matrix (ECM). Accumulated ECM destroys liver by forming fibrotic scar and subsequent nodular development ultimately leading to liver cirrhosis. Fibrotic liver contains three to ten times more ECM which in turn distorts liver parenchyma and vascular architecture and results in liver dysfunction. Hepatic Stellate Cells (HSCs) are ECM producing cells in fibrotic liver effective after activation and trans-differentiation into myofibroblasts which attains contractile, inflammatory and fibrogenic properties (Figure 1). HSCs activation results from interactions with damaged hepatocytes, Kupffer cells, disintegrated platelets and sub-fractions of leucocytes [3,4]. Progression of inflammatory and fibrogenic pathways are mediated by cytokines, genetic and epigenetic mechanisms. After chronic liver injury, fibrogenesis starts with necrosis or apoptosis of hepatocytes and inflammation connected activation of hepatic stellate cells, their trans-differentiation to myofibroblasts with enhanced expression and secretion of extracellular matrix and deposition which attains contractile, proinflammatory and fibrogenic property. In pathophysiology of liver fibrogenesis, Transforming Growth Factor-β (TGF-β), Platelet Derived Growth Factor (PDGF), endothelin-1 and Vascular Endothelial Growth Factor (VEGF) play a dominant role [4]. Genes regulating hepatocellular damage, inflammatory response to injury and Reactive Oxygen Species (ROS) generation regulates extent of hepatic damage, inflammation and ECM deposition [5]. Epigenetic mechanisms [DNA methylation, histone modifications and noncoding micro RNA (miRNA)] have been shown to orchestrate many aspects of fibrogenesis of liver [6]. Recent studies have shown that liver fibrosis is dynamic and potentially bidirectional process. Treatment aimed at underlying cause especially at early stage of the disease can reverse fibrosis to normal liver architecture by spontaneous resolution of hepatic scar. Reasons for resolution may be due to increase in collagenolytic activity, increased Matrix Metalloproteinase (MMPs) activity and decreased expression of Tissue Inhibitors of Metalloproteinase (TIMPs). Cytokine mobilization of bone marrow derived stem cells restores neutrophil function and promotes hepatic regeneration [3]. The stage at which disease become irreversible is not well established but it is believed that irreversibility attains once septal neovascularisation happens and portal pressure increases significantly [7]. Hence, we have made an attempt in this review to understand the molecular and epigenetic mechanisms involved in bidirectional liver fibrogenesis.
Pathophysiology of Bidirectional Liver Fibrosis
Natural wound healing response of liver for chronic liver injury results in the formation of hepatic scar leading to fibrosis of liver. After an acute injury, liver parenchymal cells regenerate and replace the necrotic and apoptotic cells. If hepatic injury persists, there will be failure in hepatic regeneration and substitution of hepatocytes with abundant ECM having contractile, inflammatory and fibrogenic properties [8]. Different types of cells (resident innate inflammatory cells, hepatocytes, liver sinusoidal endothelial cells and Kupffer cells) play a role in liver fibrogenesis. Activation of HSCs is a crucial step in inter-linked process of tissue injury and regeneration [9]. Quiescent HSCs present in space of Disse will be activated and trans-differentiate into myofibroblasts like cells which are responsible for ECM production and accumulation in injured liver [10]. Accumulation of ECM is due to increased synthesis and decreased degradation by over expression of TIMPs which inhibits MMPs [9]. Fibrotic liver contains three to ten times more ECM compared to normal liver includes collagen types, glycoproteins, proteoglycans and glycosaminoglycans [3]. Chief mitogen of HSCs activation is PDGF produced by Kupffer cells; macrophages are source of pro-fibrotic chemokines [11,12]. Activated HSCs activate immune response by secretion of cytokines, chemokines and interacting with immune cells. Complex network of cytokines (Table 1) modify activities of circulating immune cells, HSCs, hepatocytes, liver sinusoidal endothelial cells and Kupffer cells (Figure 2). Autocrine and paracrine secretions of cytokines activate and trans-differentiate HSCs into myofibroblasts [9]. Activated HSCs migrates to tissue repair site and secrete ECM; collagen synthesis is regulated by transcription and post-transcription. Collagen fibrils can be cross-linked by tissue transglutaminase and lysyl oxidase pathways which make collagen susceptible for collagenase activity [13]. Low density matrix is replaced by high density interstitial matrix which disturbs metabolic functions and impairs solute transport; altered cellular behavior is mediated by Integrins [11]. Damaged hepatocytes release ROS and fibrogenic mediators which stimulate inflammatory cells and fibrogenic action of activated HSCs. Activated HSCs stimulate lymphocytes by secreting inflammatory chemokines. It is a cyclic stimulation process of inflammatory and fibrogenic cells vice versa [5]. Damaged hepatocytes release inflammatory cytokines which activate Kupffer cells and stimulate the recruitment of activated T cells. This activates quiescent HSCs into fibrogenic myofibroblasts secrete cytokines. Due to chronic liver injury, activated HSCs express and deposits ECM leads fibrosis of liver. ECM degradation is inhibited by the actions of TIMPs. When etiology of liver fibrosis removed, there will be spontaneous resolution of fibrosis by apoptosis of activated HSCs and regeneration of hepatocytes. Accumulated collagen is degraded by increased activity of MMPs. Spontaneous resolution of liver fibrosis is possible after successful treatment of causative agent and may take several years depending on cause and severity of the disease [5,12]. Characteristic features of liver fibrosis reversal are decreased inflammation and decreased fibrogenic cytokines, increased collagenase activity and disappearance of myofibroblast and fibrotic scar [14]. Regression of liver fibrosis consists of thinning of fibrous septa, regeneration of hepatocytes and recovery of acinal structure [15]. Reversal of liver fibrosis can be achieved by inhibition of HSCs activation, neutralization of proliferative, fibrogenic, contractile and proinflammatory response of HSCs, stimulation of HSCs apoptosis or senescence and degradation of scar matrix. Inhibition of HSCs activation and trans-differentiation into myofibroblasts can be attained by reducing oxidant stress [10]. Interferon-β (IFN-β) inactivates HSCs and decrease production of collagen I and α Smooth Muscle Actin (α-SMA) by inhibiting PDGF and TGF-β; Interferon-γ (IFN-γ) has inhibitory action on activation of HSCs [9]. Fibrillar collagens are degraded by interstitial MMPs (MMP-1, -8 and -13) which are released in pro-enzyme form and activated by cleavage of inhibitory N-terminal peptide by plasmin. Plasmin synthesis in fibrotic liver is inhibited by synthesis of plasminogen activator inhibitor-1 expressed from activated HSCs [13]. During resolution of fibrosis, MMPs activity is increased due to decreased expression of TIMPs; monocyte/macrophage lineage expresses MMPs [16]. After removal of inflammation, macrophages are differentiated into Ly6clow phenotype which produces MMP9 and MMP12 capable of matrix degradation [14]. Altered interactions between activated HSCs and ECM favor apoptosis [5,13]. Myofibroblast apoptosis is contributed by activation of death receptor mediated pathway, increased expression of pro-apoptotic proteins (p53, Bax and Bcl-2) and decreased expression of pro-survival proteins [14]. After successful removal of causative agent, HSCs undergo caspase-8/caspase-3 dependent apoptosis. Over expression of pro-apoptotic proteins leads to caspase-9 mediated programmed cell death. Over expression of CXCL9 by macrophages and VEGF expression accelerate fibrosis resolution by angiogenesis [16].
Genetics of Bidirectional Liver Fibrosis
Genetics of liver fibrosis progression are highly complex and influenced by multiple factors. Hepatocellular apoptosis/ necrosis genes viz Bcl-xL, Fas influence the extent of hepatic damage and fibrinogenesis. Inflammatory genes viz IL-1 β, IL-6, IL-10, IFN-γ, SOCS-1 and osteopontin determines the fibrogenic response to injury. Genes regulating ROS generation (NADPH oxidase) regulate inflammation and ECM deposition [5]. Trans-differentiation of activated HSCs into myofibroblasts is mediated by down regulation of lipogenic genes like peroxisome proliferator-activated receptor gamma (PPARγ) and up-regulation of fibrogenic genes. Activated HSCs and myofibroblasts migrate to site of injury and express fibrogenic genes viz vimentin, collagen α1 (Colla1) and α-SMA stimulated by increased levels of PDGF and TGF-β [17,18]. Fibrogenic growth factors, vasoactive substances and adipokines are required for fibrogenesis (Table 2). In fibrosis resolution, expression of fibrogenic genes is decreased by inactivated HSCs in association with increased expression of genes like PPAR-γ. Genes viz GFAP, Adiporl, Adpf and Dbp are not expressed by inactive HSCs shows the difference between quiescent HSCs and inactive HSCs [19]. Gene polymorphisms play a major role in progression of liver fibrosis due to chronic liver diseases. In Alcoholic Liver Disease (ALD), polymorphisms of genes encoding alcohol metabolizing enzymes and proteins (alcohol dehydrogenase, aldehyde dehydrogenase and P450), genes encoding inflammatory mediators and antioxidants influence fibrogenesis [5]. In nonalcoholic steatohepatitis, genotypes of IL-10-1082G/ A and TNF-α 308G/ A express elevated levels of inflammatory cytokines [20]. In non-alcoholic fatty liver disease, CDKN1A variant rs762623 is related to development of liver fibrosis [21]. In primary biliary cholangitis, polymorphisms of genes encoding IL-1β, IL-1 and TNF-α are responsible for diseases progression. In Hepatitis C Virus (HCV) infection, polymorphisms of genes involved in immune response (transporter associated with antigen processing 2, specific HLA-II alleles), fibrogenic agonists (angiotensinogen and TGF-β) enhances fibrosis [5].
Epigenetics of Bidirectional Liver Fibrosis
Cellular composition and phenotype changes in chronic liver diseases are under the control of chromatin configuration of regulatory genes directed by epigenetic mechanisms [6]. Multifactorial causes influence the epigenetic mechanisms through SNPs [3]. Epigenetics are reversible changes in gene expression which are inherited through cell division without altering underlying DNA sequence; DNA methylation, post transcriptional modifications of amino acid tails of histones and non-coding RNA mediated gene silencing. These mechanisms organize many aspects of liver fibrosis by regulating chromatin structure, modifications and initiation of transcription that alters the accessibility of genes [22]. Diverse biological functions of liver are regulated by noncoding small microRNAs and play role in pathophysiology of liver [23]. Epigenetic mechanisms are determinants of gene expression during HSCs activation and deactivation by controlling transcription activity during fibrosis progression and regression. Unlike genetic mutations, epigenetic changes undergo reversion with the resolution of liver fibrosis and can be modulated pharmacologically [6,22].
DNA Methylation
Development of liver fibrosis is associated with aberrant DNA methylation patterns which lead to inappropriate gene expression. DNA methylation is regulated by DNA methyltransferases (DNMT1, DNMT3a and DNMT3b), hydroxymethylases which increases in fibrotic liver while hepatic expression of Ten Eleven Translocation (TET) demethylase is down regulated [6]. Hypermethylation of cell cycle genes (p15 and p16), tumor suppressor genes (RASSF1A and E-cadherin) and anti-fibrotic gene PPARγ is associated with liver fibrosis progression [22]. Transdifferentiation of HSCs expresses methyl-CpG-binding domain nuclear proteins (MeCP2, MBD1, MBD2 and MBD3) which are transcriptional repressors of epigenetic silencing of PPARγ gene. MeCP2 has positive regulation on expression of histone methyltransferase ASH1 which is required for expression of pro-fibrogenic genes collagen1, TIMP1 and TGF-β1. In hepatic myofibroblasts, MeCP2 regulate gene expression by direct methyl-CpG-dependent transcription and indirect post-transcriptional mechanisms [6]. Transdifferentiation of HSCs to pro-fibrogenic myofibroblast phenotype is suppressed by DNMT inhibitor 5’-aza-deoxycytidine [24].
Histone Modifications
Liver damaging agents dysregulate chromatin structure by epigenetic mechanisms which involve action of ROS on histone modification. In post-translational modifications of histone proteins, lysine methylation or acetylation regulates liver fibrosis by perspective activation of HSCs. In activated HSCs, histone methyltransferase (H3K4 methyltransferase), ASH1 is up-regulated during progression of fibrosis and binds to promoters and 5’ end of α-SMA, collagen1, TIMP-1 and TGF-β1 which results in transcriptional activation of gene expression. H3K27 methylation leads to repression of PPARγ gene; H3K9 dimethylation results in repression of inhibitory protein IĸBα leads to up-regulation of transcription factor NFĸB which has an important role in liver fibrosis. HSCs transdifferentiation requires chromatin signature H3K27me3 by recruited PPARγ gene. Lysine acetylation of histone proteins can up-regulate expression of collagen1, TIMP-1 and TGF-β1. Histone acetylation can be inhibited which can reverse myofibroblast differentiation by Histone Acetylation (HDAC) inhibitors [6,22,25].
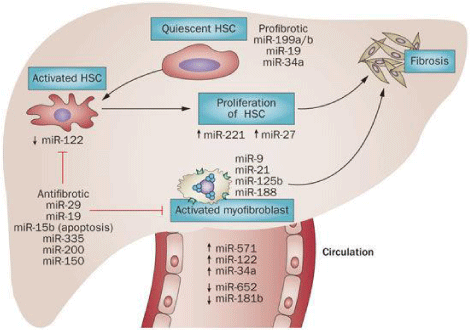
Small Non Coding RNAs (miRNAs)
miRNAs are essential for cellular process by regulating mRNA transcripts and are involved in activation of HSCs and liver fibrosis through regulation of proliferation and apoptosis (Table 3). Liver homeostasis is regulated by miRNA-122 affects on various genes involved in metabolism. miRNA-155 involved in innate and adaptive immune response by targeting TNF and promotes liver inflammation; miRNA-146a is a negative regulator of Toll Like Receptor (TLR) signaling proinflammatory response. Hepatocyte proliferation is regulated by miRNA-21 gene mediated cell cycle and DNA synthesis. Cytokines and growth factors of liver fibrosis regulate expression of pro-fibrogenic and anti-fibrogenic miRNAs (Figure 3). Key factors of fibrogenesis viz Col1α1, TGF-β receptor II, hepatocyte nuclear factor 4 α (HNF4 α) are regulated by miRNAs effect on mRNA 3’-UTR [6,22,23].
Long Non-Coding Rnas (lncRNAs) and Circular RNAs (circRNAs)
lncRNAs (exonic, intronic, overlapping and intergenic) effect gene expression by modulation of chromatin remodeling, control of gene transcription, post-transcriptional mRNA processing, protein function or localization and intracellular signaling; H19 and XIST were first identified lncRNAs for liver diseases [26]. lncRNA maternally expressed gene3 (lncRNA MEG3) located on chromosome 14q32.3 is a tumor suppressor gene which is down regulated due to TGF-β mediated methylation in disease progression of liver fibrosis [27]. Epigenetic regulation of MEG3 regulates fibrosis by inducing apoptosis by Bax/ Bcl-2 and cytoplasmic cytochrome C expressed p53 mediation. ECM synthesis will be reduced by over expression of MEG3 by suppressing cell proliferation [26]. LALR1 (human ortholog hLALR1) enhances hepatocyte proliferation through activation of Wnt/ β-catenin signaling and suppressing Axin1 [28]. Circular RNAs (circRNAs) a class of endogenous RNAs regulate gene expression at transcriptional or post-transcriptional level by acting as miRNAs sponges [29]. In bidirectional liver fibrosis, has_circ-0004018 transcribed by SMYD4 has lower expression in disease progression [30]. circHIPK3 derived from Exon2 of HIPK3 gene is a modulator of cell proliferation and significantly up-regulated in liver cancer [29].
Conclusion
Natural wound healing response to chronic liver insults results in the formation of liver fibrosis which is mediated by complex network of cytokines, growth factors, genetic and epigenetic mechanisms. Recent studies have shown that liver fibrosis is potentially bidirectional. Molecular mechanisms for liver regression in humans need to be more comprehensively defined. At which point, liver fibrosis will become irreversible is not well established, early stages may give witness for reversibility. Even though reversible epigenetic mechanisms of liver fibrosis can be modulated pharmacologically, it needs extensive research to improve anti-fibrotic drug therapies.
- Hong H, Tong W. Emerging efforts for discovering new biomarkers of liver disease and hepatotoxicity. Biomark Med. 2014; 8: 143-146. https://goo.gl/2Ya7ZT
- Jun LC, Michael P, Ahmad M, John DR. Non-invasive markers of liver fibrosis: Adjuncts or alternatives to liver biopsy? Front Pharmacol. 2016; 7: 159. https://goo.gl/ns4em2
- Krishna SN, Shashidhar KN, Lakshmaiah V, Muninarayana C. Liver fibrosis: A compilation on the biomarkers status and their significance in disease progress. Future Sci OA. 2017; 4: https://goo.gl/bfrr2x
- Axel MG, Chun-Fang G, and Olav AG. Non-invasive biomarkers for monitoring the fibrogenic process in liver: A short survey. World J Gastroenterol. 2009; 15: 2433-2440. https://goo.gl/6HyasN
- Ramón B, David AB. Liver fibrosis. J Clin Invest. 2005; 115: 209-218. https://goo.gl/1kRus8
- Eva Moran-Salvador, Jelena M. Epigenetics and liver fibrosis. Cell Mol Gastroenterol Hepatol. 2017; 4: 125-134. https://goo.gl/LxzrDf
- Sohrabpour AA, Mohamadnejad M, Malekzadeh R. Review article: the reversibility of cirrhosis. Aliment Pharmacol Ther. 2012; 36: 824-832. https://goo.gl/4W6i63
- Gressner OA, Weiskirchen R, Gressner AM. Evolving concepts of liver fibrogenesis provide new diagnostic and therapeutic options. Comp Hepatol. 2007; 6: https://goo.gl/A2Epjo
- Ebrahimi H, Naderian M, Sohrabpour AA. New concepts on pathogenesis and diagnosis of liver fibrosis; A review artcle. Middle East J Dig Dis. 2016; 8: 166-178. https://goo.gl/yXUAT5
- Scott LF. Hepatic Fibrosis. In: Schiff’s Diseases of the Liver (Edition 11). Eugene R Schiff, Michael F Sorrell, Willis C Maddrey (Ed). Wiley-Blackwell, Singapore. 2012: 297-305.
- Meena BB, Scott LF. Hepatic Fibrogenesis. In: Sherlock’s Diseases of the Liver and Biliary System. (Edition 11). James S. Dooley, Anna S.F. Lok, Andrew K. Burroughs, E. Jenny Heathcote (Ed). Wiley Blackwell, Oxford. 2011: 94-101.
- David AB. Reversibility of liver fibrosis. Gastroenterol Hepatol (N Y). 2013; 9: 737-739. https://goo.gl/WzsfK8
- Manoj KS, Sarin K. Is cirrhosis of the liver reversible? The Indian Journal of Pediatrics. 2007; 74: 393-399. https://goo.gl/M1nFNf
- Antonella P, Prakash R, and John P I. Reversibility of liver fibrosis. Fibrogenesis Tissue Repair. 2012; 5: 26. https://goo.gl/MjwccN
- Shogo O, Haruka H, Kazuhiko W, Katsuhiko H, Kenya K, and Masahiko Y. Natural regression of fibrosis in chronic hepatitis B. World J Gastroenterol. 2016; 22: 5459-5466. https://goo.gl/p5nSy5
- Seki E, Brenner DA. Recent advancement of molecular mechanisms of liver fibrosis. J Hepatobiliary Pancreat Sci. 2015; 22: 512-518. https://goo.gl/WPCYs4
- Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008; 134: 657-667. https://goo.gl/RTVEee
- Hernández-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel M, Yue Z, et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012; 142: 938-946. https://goo.gl/GEG5Lh
- Kisseleva T, Cong M, Paik Y, Scholten D, Jiang C, Benner C, et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc Natl Acad Sci USA. 2012; 109: 9448-9453. https://goo.gl/5JhkU3
- Ioana CB, Mircea VM, Raluca MP, Stefan CV, Lorena C, Daniela MM, et al. Cytokines genotype-phenotype correlation in nonalcoolic steatohepatitis. Oxidative Medicine and Cellular Longevity. 2017; 7. https://goo.gl/ZP2Cqy
- Aravinthan A, Mells G, Allison M, Leathart J, Kotronen A, Yki-Jarvinen H, et al. Gene polymorphisms of cellular senescence marker p21 and disease progression in non-alcohol-related fatty liver disease. Cell Cycle. 2014; 13: 1489-1494. https://goo.gl/na9YBE
- Atta HM. Reversibility and heritability of liver fibrosis: Implications for research and therapy. World J Gastroenterol. 2015; 21: 5138-5148. https://goo.gl/ECGvgM
- Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013; 10: 542-552. https://goo.gl/9MFQNU
- Mann J, Oakley F, Akiboye F, Elsharkawy A, Thorne AW, Mann DA. Regulation of myofibroblast transdifferentiation by DNA methylation and MeCp2: implications for wound healing and fibrogenesis. Cell Death Differ. 2007; 14: 275-285. https://goo.gl/QiycQd
- Perugorria MJ, Wilson CL, Zeybel M, Walsh M, Amin S, Robinson S, et al. Histone methyltransferase ASH1 orchestrates fibrogenic gene transcription during myofibroblast transdifferentiation. Hepatology. 2012; 56: 1129-1139. https://goo.gl/bkNhpz
- Takahashi K, Yan I, Haga H, Patel T. Long non-coding RNA in liver diseases. Hepatology. 2014; 60: 744-753. https://goo.gl/Aj1wcu
- Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S et al. MicroRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011; 30: 4750-4756. https://goo.gl/6y4akq
- Xu D, Yang F, Yuan JH, Zhang L, Bi HS, Zhou CC et al. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating wnt/beta-catenin signaling. Hepatology. 2013; 58: 739-751. https://goo.gl/zsuiBR
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016; 7: 11215. https://goo.gl/uVCK6c
- Liyun Fu, Ting Yao, Qingqing Chen, Xiaoyan Mo, Yaoren Hu and Junming Guo. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017; 8: 58405-58416. https://goo.gl/6NhnjA

Sign up for Article Alerts