Real-Time Polymerase Chain Reaction Coupled With High-Resolution Melting Analysis for the Differential Detection of Mycobacterium Avium Subspecies Avium versus Mycobacterium Avium< Subspecies Paratuberculosis in Animal Lesion Specimens
Worasak Kaewkong1*, Gunticha Suwanmanee1, Phattarin Pothipan1, Tarid Purisotayo2 and Seksit Singjam3
1Faculty of Medical Science, Naresuan University, Phitsanulok 65000, Thailand2Faculty of Veterinary Science, Mahasarakham University, Maha Sarakham 44150, Thailand
3Veterinary Research and Development Centre-Lower Northern Region, Phitsanulok 65000, Thailand
*Address for Correspondence: Worasak Kaewkong, Faculty of Medical Science, Naresuan University, Phitsanulok, 65000 Thailand, Tel: +668-671-513-13; ORCID ID: orcid.org/0000-0002-7130-5827; E-mail: worasakk@nu.ac.th; worasak_bc@hotmail.com
Submitted: 29 May 2019; Approved: 08 June 2019; Published: 11 June 2019
Citation this article: Kaewkong W, Suwanmanee G, Pothipan P, Purisotayo T, Singjam S. Real-Time Polymerase Chain Reaction Coupled With High-Resolution Melting Analysis for the Differential Detection of Mycobacterium avium Subspecies avium versus Mycobacterium avium Subspecies Paratuberculosis in Animal Lesion Specimens. American J Current App Res Microbiol. 2019;1(1): 017-021.
Copyright: © 2019 Kaewkong W, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Keywords: Duck; High resolution melting analysis; Mycobacterium avium; Subspecies
Download Fulltext PDF
Two main animal pathogenic subspecies of Mycobacterium avium are M. avium avium (Maa) and M. avium Paratuberculosis (Map). Their pathogenicity is host-specific, Maa causing avian tuberculosis in poultry whereas Map commonly cross-infects to ruminant. Veterinary diagnosis of M. avium infections is microscopic examination of acid-fast bacilli or culture in Löwenstein-Jensen medium, which are time-consuming and low sensitivity. This present study aimed to apply real-time PCR coupled with High-Resolution Melting (HRM) analysis for differential detection of Maa in Thai domestic ducks. Specific primer targeting host-expression dependent (hed) region was designed, PCR product of Maa were amplified from duck’s tissue lesions whereas Map were amplified from cow and deer. HRM real-time PCR was performed and analyzed. Different HRM patterns were showed and melting temperature were analyzed at 83.26 ± 0.12°C for Maa and 84.04 ± 0.09°C for Map. This technique can detect as few as 102 DNA copies and present high specificity by negative amplification of other pathogenic bacterial species. This technique is sensitive, specific, rapid and does not require fluorescent probes or post-PCR electrophoresis. Our technique is a possible new tool for the detection of Maa and Map infection in tissue specimens.
Introduction
Mycobacterium avium was recognised as a cause of tuberculosis primarily in chickens, ducks and other fowls, but can also infect to an extensive range of different animal species [1]. At least 20 different types of M. avium have been identified which cause tuberculosis in most avian species [2]. Avian tuberculosis generally is transmitted by direct contact with infected birds and can susceptible to human and bovine after ingestion of contaminated feed and water. All avian species are susceptible to infection by M. avium. Humans, most livestock species, and other mammals can also become infected. The susceptibility is high in domestic fowl (Gallus domesticus), pheasants, wild birds, swine, rabbit and human with immunocompromised [3]. In human, there are many authenticated cases of M. avium infection although humans are considered highly resistant to this pathogenic species. Infection is more likely to occur in persons with pre-existent diseases, especially those involving the lungs, and in immune systems impaired-person [4].
Infection with M. avium causes serious problems in tropical countries in Asia particularly Thailand where a number of infected-poultry including chickens and ducks was reported. There are two main pathogenic subspecies of M. avium were recognized as an important problem are M. avium subsp. avium (Maa) and M. avium subsp. Paratuberculosis (Map).
Several regions in Thailand were presented the overlapping of these two subspecies infections. Avian tuberculosis is one of the most important diseases that affect both domestic and pet birds, the disease is most often caused by Mycobacterium avium belonging Maa whereas Map is the etiologic agent of Johne’s disease causing chronic diarrhea, malnutrition, and muscular wasting in ruminants [5,6]. Maa and MAP are closely related and similar by morphology or genetics information, and both can infect to the human in the term of M. avium Complex (MAC). Notably, MAC comprising M. avium subsp. avium, M. avium subsp. Paratuberculosis and M. avium subsp. Silvaticum or even M. intracellulare, may also infect different animal species like cattle, deer, horses, swine, and exotic species besides causing infection in immunocompromised human [7].
Microbiological methods based on microscopic examination of Acid-Fast Bacilli (AFB) presented the low sensitivity regarding to staining procedures and proficiency of technician. Regularly, the bacterial culture in Lowenstein–Jensen medium is specific to M. avium but it’s time consuming (6-8 weeks). They are some definite weak points from routine detection protocol, the stained AFB as well as culture colonies of Maa and Map cannot distinguish each other. Furthermore, they have low sensitivity, especially in early or lightly infected cases.
High Resolution Melting (HRM) analysis of DNA is a simple solution for genotyping, scanning of mutation and other aspects on sequence analysis. The melting profile of DNA, a product from PCR amplification, depends on GC content, length, and which is monitored with saturating dyes that fluorescent in the presence of double-stranded DNA [8,9]. In the field of veterinary and animal science, HRM has been used for detection or differentiation of various pathogenic microorganisms for example, in viruses, bacteria, or parasites [10-12]. For mycobacterial, HRM analysis was applied in many aspects, for example, to differential identification at the species level in clinical isolates or in laboratory standard strains from American Type Culture Collection (ATCC) and the Culture Collection [13,14].
For the studies in Mycobacterium avium such as HRM was applied for identifying the polymorphisms in Map types I, II, and III [15], for sub-typing Map by analysis of Short Sequence Repeats (SSR) loci [16], differentiating Map cattle- and sheep-type [17], and genotyping the members of M. avium-Intracellulare (MAI) complex [18]. However, real-time Polymerase Chain Reaction (real-time PCR) with HRM for differentiation of Maa and Map in animal lesion specimens has not been reported yet. In the present study, we developed a real-time PCR coupled with HRM to differential detection of Maa and Map in animal lesion specimens including poultry (ducks) and ruminant (cow and deer) hosts. In addition, we also validate the diagnostic specificity and sensitivity of this approach.
Materials and Methods
Sample collection and research ethic
The animal experiments and related-protocols in this study were approved, the authors were certified by the Institutional Animal Care and Use Committee (Thai-IACUC) cooperated with the Institute of Animal for Scientific Purposes Development (IAD) of Thailand (permit number. U1-05294-2559) and Ethical Principle for the Use and Care of Animals in Scientific Research (ACUC 5/2560), based on the Ethics of Animal Experimentation of the National Research Council of Thailand.
Forty-one DNA samples of M. avium infected-domestic ducks were obtained from Veterinary Research and Development Centre (VRDC)-Lower Northern Region, Phitsanulok, Thailand which they were portions of the leftover specimens received from routine laboratory diagnosis. Among 41 DNA samples from ducks, they were isolated from lesions including wings (n = 19), lungs (n = 9), intestines (n = 11) and kidneys (n = 2). Two DNA samples of M. avium infected cow and deer were kindly provided from National Institute of Animal Health of Thailand which were recorded as isolated from the lesions at lungs of both infected animals. DNA samples of Pseudomonas aeruginosa, Enterococcus faecalis, Escherichia coli, Staphylococcus aureus, Pasteurella multocida type a, Salmonella Group B (derived from microbiology laboratory of VRDC-Lower Northern Region). For negative control, three DNA samples from non-infected ducks and two DNA samples from non-infected cow were used. All DNA samples were stored at -80°C until usedDNA amplification by Polymerase Chain Reaction (PCR)
For specific DNA amplification, a pair of specific primers were designed to targeting the host-expression dependent (hed) region of Maa (GenBank accession number: AJ011837) and Map (GenBank accession number: AJ011838). For the PCR amplification, forward primer was Mav100-F (5’- ATG AAA GCC ATA CCC GAC GTC CCT -3’) and reverse primer was Mav100-R (5’- TGC GGT ACT CGA TCA TGC TGT CCT -3’). Platinum® Taq DNA Polymerase (Invitrogen, Carlsbad, CA, USA) was used for PCR amplification by the conventional PCR. The total reaction volume was 25 µl with 10x PCR buffer, 50 mM MgCl2, 10 mM dNTP mix, 10 µM of forward and reverse primers, and Platinum® Taq DNA Polymerase. Using the PCR conditions of 1 cycle for pre-incubation at 95°C for 5 min, 30 cycles of amplification step at 95°C for 30 sec, 57°C for 30 sec, and 72°C for 30 sec.
Cloning and sequencing of control DNA
For DNA cloning, sequencing and construction of control plasmids, Positive control plasmids of Maa and Map were constructed by cloning of the relevant PCR products into the pCR®2.1-TOPO® vector (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions and were subsequently propagated in Escherichia coli DH5-alpha strain (Real Biotech Corp., Taipei, Taiwan). After amplification and agarose gel electrophoresis, the bands of PCR product were purified by HiYield™ Gel/PCR DNA Mini Kit, (Real Biotech Corp., Taipei, Taiwan) and the nucleotide sequence were determined by 3500 Genetic Analyzer (Applied Biosystems, Inc. [Hitachi Ltd], Tokyo, Japan).
High-resolution melting analysis
For Real-time PCR and HRM analysis, LightCycler ® 96 Real-Time PCR System for detection and analysis were used for amplification and quantification. A LightCycler® 480 High Resolution Melting Master (Roche Applied Science, Mannheim, Germany) was used as recommended by the manufacturer, including Master Mix (FastStart Taq DNA Polymerase, reaction buffer, dNTP mix and HRM dye), MgCl2 and H2O PCR-grade. The PCR mixture was prepared to contain 1x HRM Master, 2.2 mM MgCl2, 0.8 µM of each 2 primers (Mav100-F and Mav100-R). The total reaction volume was 20 µl. The PCR cycling for HRM curve acquisition was run under the following conditions: 1 cycle for pre-incubation at 95°C for 10 min, 45 cycles of amplification step at 95°C for 10 sec, 57°C for 8 sec, and 72°C for 15 sec. After amplification, the PCR products were melted by raising the temperature from 60°C to 95°C, with an increment of 0.1°C/sec. After, the amplification curve was monitored real-time, the melting temperatures (Tm) and melting curve were determined and analyzed.
Evaluation of sensitivity and specificity of detection
For determination of analytical sensitivity, the ten-folded serial dilution from 109 to 1 copies of Maa and Map positive control plasmids were prepared. For determination of analytical specificity, DNA samples of P. aeruginosa, E. faecalis, Escherichia coli, Staphylococcus aureus, Pasteurella multocida type a, Salmonella Group B were used. They were subsequently analysed by Real-time PCR and HRM analysis as described above. The diagnostic values were calculated using standard methods [19].
Results
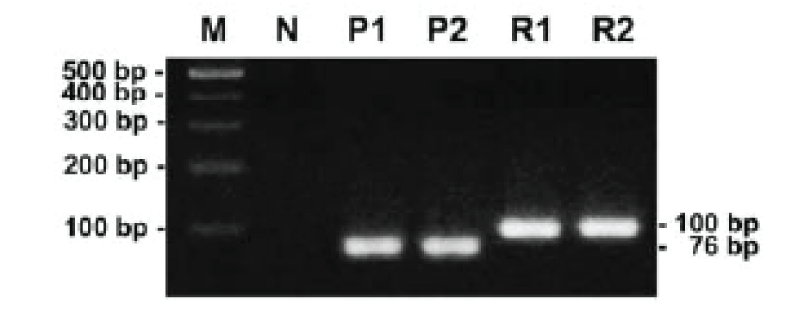
For PCR amplification and sequencing analysis of M. avium DNA. Using the primer targeting the region that described in material and methods, we successfully amplified a predicted 76 bp and 100 bp product specific from DNA of Maa and Map, respectively (Figure 1).
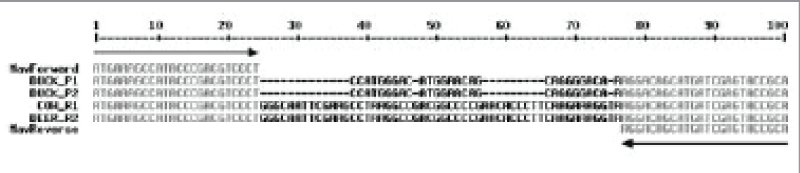
After purification and DNA sequencing, the obtained sequences were matched to the sequences of genes from which the primers were designed. There are some deletions occur in the region of Maa DNA that amplified from duck’s infected-tissue compared with 100 bp of Map DNA from infected tissue of the cow and the deer (Figure 2).
For HRM real-time PCR for differential detection of Maa and Map, the method yielded the positive results for all 41 Maa-infected and 2 Map-infected DNA samples. After amplification, the melting temperatures (Tm) and melting curve were analysed. Representative melting temperatures were 83.26 ± 0.12°C for Maa and 84.04 ± 0.09°C for Map (Figure 3A-C). DNA from Maa and Map could be obviously distinguished by the specific melting temperatures determined by the difference plots (Figure 3A), the normalized melting curves (Figure 3B), and the melting peaks (Figure 3C).
To standardization of the HRM real-time PCR, there is no fluorescent signal were detected when evaluated with the no DNA control of each experiments, especially in 3 DNA samples from non-infected ducks and 2 DNA samples from non-infected cow. For analytical specificity of HRM real-time PCR, there are no positive result from amplicon at specific melting temperatures was perceived when various DNAs from other pathogenic bacterial species were analysed which demonstrates the specificity of this method (Figure 3D). In addition, the analytical sensitivity was determined using ten-fold serial dilutions (109 - 1 copies) of the Maa and Map positive control plasmids. The sensitivity was illustrated by DNA of both Maa and Map could be detected at the lowest concentration used (102 copies) when considering 45 cycles as the cut-off detection limit (Figure 3E-F).
Discussions
This is the report of HRM real-time PCR to distinguish between Maa and Map in DNA samples extracted from tissue lesion specimens of infected animals in a single assay. We present the application for homogenous genotyping of M. avium subspecies without fluorescent labelled-probe and post-PCR electrophoresis. Therefore, the method consequently reducing the expense on a cost per sample and the time consuming. Simplification of interpretation is obtained by different sequences from different subspecies represent a change in the temperature in analysed-melting peak. The HRM real-time PCR offers a new alternative for rapid, sensitive and subspecies specific for differentiation and detection of two main pathogenic M. avium subspecies in poultry and ruminant hosts. The high sensitivity and specificity suggest the present test will be of diagnostic value. The analytical sensitivity in this study was only 100 copies of Maa and Map DNA. Moreover, real-time PCR with HRM produced no cross reaction between Maa and Map or various DNAs from other bacterial species that usually infected duck and cow indicating high specificity.
There are several studies mention the application of HRM real-time PCR for mycobacterial. Recently in 2017, the researchers defined comparison of melting curves of different non-tuberculosis mycobacteria and presented the melting curves corresponding to the 6 unique-specific HRM groups (M. tuberculosis complex, M. kansasii, M. simiae, M. fortuitum, M. abscessus–M. chelonae group, and M. avium complex) for the mycobacterial rpoBC locus in several collected clinical specimens [13].
Previously, the standardization of HRM real-time PCR were verified by laboratory standard of mycobacterial strains [14]. In that study the DNA were amplified by primer targeting 16S rRNA gene, the Tm values for M. tuberculosis and M. bovis of the were 86.00 ± 0.00°C. M. smegmatis, M. ulcerans and M. xenopi, the Tm values were 86.40 ± 0.00°C, 86.20 ± 0.00°C and 86.60 ± 0.00°C, respectively. The Tm values for M. avium, M. avium subspecies Paratuberculosis and M. intracellulare of the M. avium Complex (MAC) were 86.20 ± 0.00°C, 86.60 ± 0.00°C and 86.80 ± 0.00°C, respectively. In our present study, we use the primer targeting at hed region and the Tm values at 83.26 ± 0.12°C for Maa and 84.04 ± 0.09°C for Map were obtained. We presented practically almost 1°C for distinguish of Maa and Map whereas previous study is only 0.4°C. In addition, DNA samples of the related pathogenic bacterial species in this group of animals, the test did not give rise to an identifiable Tm or melting peak of Maa and Map, indicating the specificity of this method.
Regarding to the samples those sent from local farmers were infected animals with the lesions were appeared, the technique optimized for detection of the infection by lesion samples should be appropriated. However, even this study attempted to apply the techniques for detection of Mycobacterium avium infection in lesion specimens, but further verifying by blood or another animal specimen was very challenged. In addition, because the primer was designed to target the sequence of Maa and Map directly. Therefore, the technique can be applied to detect these Mycobacterium avium subspecies infection in other animals or even in human.
In conclusion, this study highlights a HRM real-time PCR for the differentiation of the 2 main pathogenic subspecies of M. avium, Maa and Map. We successfully developed the rapid method for this approach, especially appropriates for the DNA samples extracted from M. avium infected lesion tissues from animal. This HRM real-time PCR can potentially be used for subspecies-specific epidemiological surveys of M. avium in domestic livestock, especially in Thailand where M. avium infections are usual informed.
Acknowledgments
This study was financially supported by the grants from Naresuan University (grant number: R2558C034) and the National Research Council of Thailand (grant number: R2559B066).
- Thorel MF, Huchzermeyer H, Weiss R, Fontaine JJ. Mycobacterium avium infections in animals. Literature review. Vet Res. 1997; 28: 439-447. https://bit.ly/2F3rpMZ
- Thorel MF, Krichevsky M, Levy-Frebault VV. Numerical taxonomy of mycobactin-dependent mycobacteria, amended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. Paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum subsp. nov. Int J Syst Bacteriol.1990; 40: 254-260. https://bit.ly/2MzZD0v
- Dhama K, Mahendran M, Tiwari R, Singh SD, Kumar D, Singh S, et al. Tuberculosis in birds: Insights into the Mycobacterium avium infections. Vet Med Int. 2011; 2011: 712369. https://bit.ly/2MDg2Br
- Ramirez J, Mason C, Ali J, Lopez FA. Mycobacterium avium complex pulmonary disease: management options in HIV-negative patient. J La State Med Soc. 2008; 160: 248-254. https://bit.ly/2R4DUwq
- Mijs W, De Haas P, Rossau R, van Der Laan T, Rigouts L, Portaels F, et al. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and M. avium subsp. hominissuis for the human/porcine type of M. avium. Int J Syst Evol Microbiol. 2002; 52: 1505-1518. https://bit.ly/2Zlful5
- Rathnaiah G, Zinniel DK, Bannantine JP, Stabel JR, Gröhn YT, Collins MT, et al. Pathogenesis, molecular genetics, and genomics of Mycobacterium avium subsp. Paratuberculosis, the etiologic agent of Johne’s disease. Front Vet Sci. 2017; 4: 187. https://bit.ly/2ZgkTdh
- Thorel MF, Huchzermeyer H, Michel AL. Mycobacterium avium and M. intracellulare infection in mammals. Rev Sci Tech. 2001; 20: 204-218. https://bit.ly/2Wy39sl
- Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics.2007; 8: 597-608. https://bit.ly/2Ka90lG
- Tong SY, Giffard PM. Microbiological applications of high-resolution melting analysis. J Clin Microbiol. 2012; 50: 3418-3421. https://bit.ly/2WrJXMJ
- Chamings A, Hewson KA, O’Rourke D, Ignjatovic J, Noormohammadi AH. High-resolution melt curve analysis to confirm the presence of co-circulating isolates of avian nephritis virus in commercial chicken flocks. Avian Pathol.2015; 44: 443-451. https://bit.ly/2XyQoPu
- Miller M, Zorn J, Brielmeier M. High-resolution melting curve analysis for identification of Pasteurellaceae species in experimental animal facilities. PLoS One. 2015; 10: e0142560. https://bit.ly/2MG8ldP
- Santos GB, Espinola SM, Ferreira HB, Margis R, Zaha A. Rapid detection of Echinococcus species by a high-resolution melting (HRM) approach. Parasit Vectors. 2013; 6: 327. https://bit.ly/2I7DLFO
- Khosrvai AD, Hashemzadeh M, Hashemi-Shahraki A, Teimoori A. Differential identification of mycobacterial subspecies using high-resolution melting analysis. Front Microbiol. 2017; 23: 2045. https://bit.ly/2wOcWA2
- Issa R, Abdul H, Hashim SH, Seradja VH, Shaili N, Hassan NA. High resolution melting analysis for the differentiation of Mycobacterium species. J Med Microbiol. 2014; 63: 1284-1287. https://bit.ly/2WBkLZ2
- Castellanos E, Aranaz A, De Buck J. PCR amplification and high-resolution melting curve analysis as a rapid diagnostic method for genotyping members of the Mycobacterium avium–intracellulare complex. Clin Microbiol Infect. 2010; 16: 1658-1662. https://bit.ly/2WtIXI3
- Ricchi M, Barbieri G, Cammi G, Garbarino CA, Arrigoni N. High-resolution melting for analysis of short sequence repeats in Mycobacterium avium subsp. Paratuberculosis. FEMS Microbiol Lett. 2011; 323: 151-154. https://bit.ly/2R5dpXV
- Douarre PE, Cashman W, Buckley J, Coffey A, O’Mahony JM. High resolution melting PCR to differentiate Mycobacterium avium subsp. Paratuberculosis cattle type and sheep type. J Microbiol Methods. 2012; 88: 172-174. https://bit.ly/2R1VJML
- Castellanos E, Aranaz A, De Buck J. Rapid identification and differentiation of Mycobacterium avium subspecies Paratuberculosis types by use of real-time PCR and high-resolution melt analysis of the MAP1506 locus. J Clin Microbiol. 2010; 48: 1474-1477. https://bit.ly/2R3FCyh
- Galen RS. Predictive value and efficiency of laboratory testing. Pediatr Clin North Am. 1980; 27: 861-869. https://bit.ly/2XJ5u56


Sign up for Article Alerts